
Plants produce and build up triacylglycerol (TAG, familiar to most people as vegetable oils) in their seeds as a main carbon and energy resource for seedling development. Plant seed oils are important for both human diet and renewable feedstock of industrial applications.
Plant oils are energy-rich — the energy content of TAG is much higher than carbohydrates and proteins. Vegetable oils are also widely used for non-food purposes, e.g. the production of soaps, detergents, shampoo, and lubricants. Global demand for plant oils is increasing rapidly and estimated to be doubled by 2030. In order to meet the growing demand, it is necessary to increase plant oil production by expanding plant acreage or improving oil yield. Increasing oil yield in crops translates to greater economic values.
WRINKLED1 (WRI1), an APETALA2 (AP2) transcription factor, is an essential regulator of plant oil biosynthesis [1]. The Arabidopsis WRI1 loss-of-function mutant, wri1-1, was first identified by Prof. Christoph Benning’s group through mutant screening [2]. The wri1-1 seeds displays “wrinkled” seed coat and ~80% reduction of seed oil content compared to the wild-type. The transcriptomic comparison of the wild-type and wri1-1 plants indicates that numerous genes with reduced expression in wri1-1 encode enzymes in the glycolytic and fatty acid biosynthetic pathways [3]. Further work has validated that the genes involved in the process of glycolysis and fatty acid biosynthesis are targets of AtWRI1 [4,5]. WRI1 is hence considered as a transcriptional “master regulator” which controls plant seed oil production.
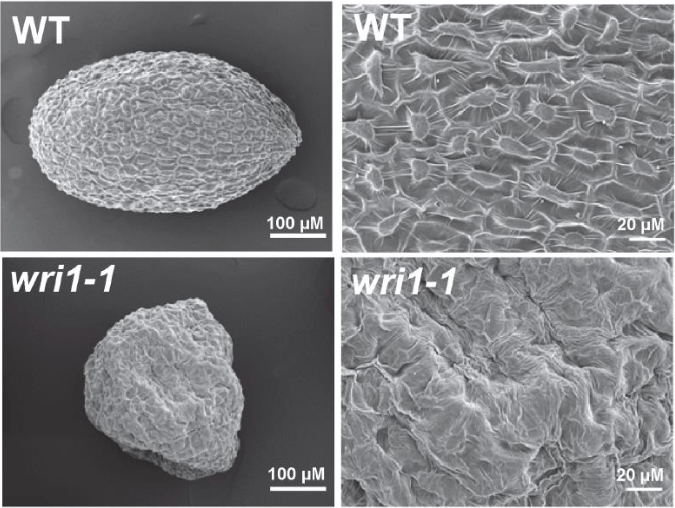
Image published with permission from PLoS from https://doi.org/10.1371/journal.pone.0068887
At the transcriptional regulation level, critical transcription factors involved in seed maturation, such as LEAFY COTYLEDON1 (LEC1) and LEAFY COTYLEDON2 (LEC2), are important regulators to activate the expression of WRI1 [4,6]. Another Arabidopsis transcription factor, FUSCA3 (FUS3) acts as an activator of WRI1 expression as well [7]. Arabidopsis transcription factor MYB89 has recently been identified to repress the expression of WRI1 [8].
Almost a decade since the discovery of AtWRI1, it remains unclear how other regulatory mechanisms, such as post-translational modifications and protein-protein interactions, influence WRI1 activity. At the protein level, functional domains (other than AP2 domains) or motifs in WRI1 are still not well characterized. In the past five years, nevertheless, progress has been made in our understanding of the mechanisms underlying the regulation of WRI1 in aspects of post-translational modifications and interacting proteins. This progress paves new routes to utilize WRI1 for bioengineering of TAG production in oil crop plants.
AtWRI1 protein degradation is perhaps a mechanism leading to down-regulation of oil biosynthesis. CULLIN3-based E3 ligase adaptor BTB/POZMATH (BPM) proteins have been identified as interacting partners that present AtWRI1 to the 26S proteasome for degradation [9]. Our recent work using multiple computational programs has identified three intrinsic disorder regions (IDRs) in the AtWRI1 protein [10]. We have also identified a PEST motif (a protein degradation signal) associated with the AtWRI1 C-terminal IDR3 and a transactivation domain (TAD) in AtWRI1.
Deletion of the IDR3-PEST motif or phosphorylation-deficient mutations in the presumed phosphorylation sites in the same motif results in enhanced protein stability. Transgenic plants expressing engineered genes encoding the IDR3-PEST-deletion or phosphorylation-deficient in the IDR3-PEST motif of AtWRI1 show elevated TAG production compared to that expressing the wild-type AtWRI1. Therefore, phosphorylation in the IDR3-PEST motif is proposed to be another regulatory mechanism that regulates WRI1 protein stability [10].
We have also identified that AtWRI1 physically interacts with 14-3-3 proteins, a family of phosphopeptide-binding proteins that are widely conserved in eukaryotes. In tobacco transient expression system, an Arabidopsis 14-3-3 enhances AtWRI1 protein stability, transcriptional activity, and AtWRI1-mediated TAG production [11]. KIN10 (an SNF1-related protein kinase) physically interacts with AtWRI1 to trigger phosphorylation, which is critical for the degradation of the AtWRI1 protein [12]. Mediator 15 (MED15), a subunit of the Arabidopsis mediator complex, also physically interacts with AtWRI1 [13].
Since the discovery of AtWRI1, numerous WRI1 orthologs have been identified in other plant species, both in dicot and monocot. The function of the WRI1s is conserved in mediating oil biosynthesis. Protein structural features and motif, such as the “VYL”[14], IDR and PEST motif [10], are common in the WRI1s [15].
The WRI1s have been successfully applied in translational research. Overexpression of AtWRI1 or WRI1 orthologs leads to elevated seed oil yields in transgenic plants [1,15,16]. Overexpression of ZmWRI1 increases kernel oil content in both greenhouse testing and field trial [17].
Much remains to be done in future studies of WRI1. Additional regulators, which mediate WRI1 expression, need to be identified. With recent identification of WRI1 functional domains and interacting partners, such as IDR, 14-3-3, and KIN10, the new focus will be on detailed characterization of the structure-function relationship. Given that plants live in a dynamic environment, future work should also focus on how the WRI1 activity is mediated in the perception of the various environmental cues.
We should not overlook the recent findings that WRI1 have target genes other than those known to be involved in glycolysis and fatty acid biosynthesis [18]. Further understanding of WRI1’s gene targets that are unrelated to oil biosynthesis is critical for the reduction of possible side effects of using WRI for bioengineering, which has been reported in some transgenic plants overexpressing WRI1 [1,16]. Approaches, such as protein engineering to enhance WRI1 binding specificity, can be considered as strategies in generating oil crops in the future.
These findings are described in the article entitled WRINKLED1 transcription factor: How much do we know about its regulatory mechanism? recently published in the journal Plant Science. This work was conducted by Que Kong from Michigan State University (USA) and Wei Ma from Nanyang Technological University (Singapore). Que Kong is currently working at Nanyang Technological University. The research project of WRI1 in the Ma lab is supported by a Nanyang Technological University Startup grant (to W.M.).
References:
- Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575-585.
- Focks N, Benning C (1998) wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91-101.
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191-1206.
- Baud S, Mendoza MS, To A, Harscoet E, Lepiniec L, et al. (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825-838.
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, et al. (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476-487.
- Casson SA, Lindsey K (2006) The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol 142: 526-541.
- Yamamoto A, Kagaya Y, Usui H, Hobo T, Takeda S, et al. (2010) Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol 51: 2031-2046.
- Li D, Jin C, Duan S, Zhu Y, Qi S, et al. (2017) MYB89 Transcription Factor Represses Seed Oil Accumulation. Plant Physiol 173: 1211-1225.
- Chen L, Lee JH, Weber H, Tohge T, Witt S, et al. (2013) Arabidopsis BPM Proteins Function as Substrate Adaptors to a CULLIN3-Based E3 Ligase to Affect Fatty Acid Metabolism in Plants. Plant Cell 25: 2253-2264.
- Ma W, Kong Q, Grix M, Mantyla JJ, Yang Y, et al. (2015) Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J 83: 864-874.
- Ma W, Kong Q, Mantyla JJ, Yang Y, Ohlrogge JB, et al. (2016) 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J 88: 228-235.
- Zhai Z, Liu H, Shanklin J (2017) Phosphorylation of WRINKLED1 by KIN10 Results in Its Proteasomal Degradation, Providing a Link between Energy Homeostasis and Lipid Biosynthesis. Plant Cell 29: 871-889.
- Kim MJ, Jang IC, Chua NH (2016) The Mediator Complex MED15 Subunit Mediates Activation of Downstream Lipid-Related Genes by the WRINKLED1 Transcription Factor. Plant Physiol 171: 1951-1964.
- Ma W, Kong Q, Arondel V, Kilaru A, Bates PD, et al. (2013) Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS One 8: e68887.
- Kong Q, Ma W (2018) WRINKLED1 transcription factor: How much do we know about its regulatory mechanism? Plant Sci 272: 153-156.
- Marchive C, Nikovics K, To A, Lepiniec L, Baud S (2014) Transcriptional regulation of fatty acid production in higher plants: molecular bases and biotechnological outcomes. Eur J Lipid Sci Technol 116 1332–1343.
- Shen B, Allen WB, Zheng P, Li C, Glassman K, et al. (2010) Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol 153: 980-987.
- Kong Q, Ma W, Yang H, Ma G, Mantyla JJ, et al. (2017) The Arabidopsis WRINKLED1 transcription factor affects auxin homeostasis in roots. J Exp Bot 68: 4627-4634.









