The cellular membrane is a biological circuit board where multiple nanostructures (e.g. proteins) process dynamic information of the extracellular environment through diverse recognition and clustering steps. Inspired by the cellular membrane, we developed a nanoparticle computing platform on a supported lipid bilayer, termed a lipid nanotablet (LNT). In the LNT, a two-dimensional supported lipid bilayer is a chemical circuit board to tether nanoparticle computing units to configure the desired logic circuit, and a fluidic membrane allows the computing units to freely diffuse on the surface and mutually interact. The nanoparticles are modified with molecular ligands (DNA in this case) that take up molecular inputs in a solution phase to encode logic gates under multiple design schemes.

Image republished with permission from Jwa-Min Nam
As a result of logic gates based on input combination, interparticle interactions, either the assembly or disassembly of a nanoparticle pair, are triggered. Such configuration change of the computing units can be identified in a real-time by monitoring the scattering signal of each particle under dark-field microscopy and the homebuilt software that allows for digitizing and quantifying particle images in a massively parallel manner.
Nanoparticle computing units are further classified into two types by its mobility: immobile Nano-Receptors and mobile Nano-Floaters. The mobility of the nanoparticle is modulated by the ratio of the number of surface tethering ligand (biotinylated DNA) to the total number of ligands on the particle surface, which affects a tethering order. Both nanoparticle types are programmed to interact only with the other type in a pairwise manner, where the Nano-Receptors offer a spatially confined reaction center, and the Nano-Floaters diffuse across the surface and carry information to the Nano-Receptor counterpart. The interaction between a pair of nanoparticles are programmed through the design schemes in two different scales, namely, interface programming and network programming by DNA designs on nanoparticles. The interface programming embeds the desired logic into computing unit interactions through ligand-level design and the combination of ligands, such as DNA hairpin ligands for assembly AND logic gate.
We demonstrated single-input YES, two-input AND, OR, and INHIBIT logic gates, as well as fan-in and fan-out modality for multiple input and output, respectively, from a single logic computation. The network programming is an approach that configures a logic circuit through connectivity of nanoparticle logic gates. Serial wiring of disassembly and assembly interactions, in which output of a disassembly logic gate is connected to the other assembly logic gate, results in AND connection of two logic gates. Together with parallel wiring of logic gates that yields an OR connection, the network programming can be modularly combined with the interface programming to design complex logic circuit as a 2-to-1 multiplexer.
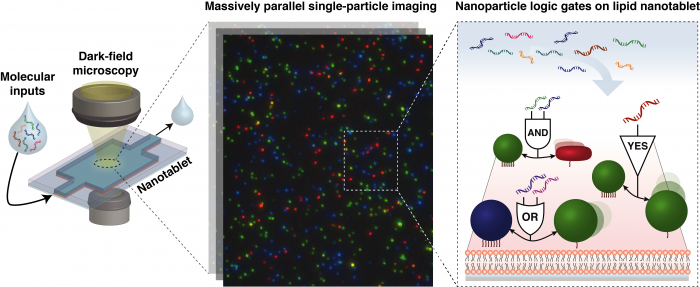
Image republished with permission from Jwa-Min Nam
The significance of the LNT concept/platform lies in the confinement of computing units to a fluidic 2D surface enables modular design and implementation of single-nanoparticle logic computations in solution phase. Unlike the solution phase, tethering computing units to the supported lipid bilayer allows for discriminating roles of computing units by their mobility and interfacial engineering between particles in a reliable manner. Using the modularity of this system, we can scale up logic circuits by connecting simple logic gates, which is difficult to achieve with other nanoparticle computing methods.
Along with the controllability achieved through the spatial restriction, the reduction in dimensionality gives scalability to the system through wiring and parallelism of signal detections for the particles on lipids both with logic implementation and in spatially distributed computing units. The LNT will be the key concept/platform to build autonomous nanosystems in molecular diagnostics and smart sensors that have unprecedented detection signal analyzing capability for a myriad of different target molecules and can be plugged into smart cell-interfacing systems.








