In situ hybridization is a common laboratory technique, widely employed to visualize gene expression or chromosomal location of DNA sequences. It is a very powerful tool, which allows combining the information about a gene with the spatial information, identifying the tissues or cells where it is acting. Recently, our groups at the Villefranche-sur-mer Developmental Biology Laboratory (France) and the Sars International Centre for Marine Molecular Biology (Bergen, Norway) developed an alternative protocol, which produced sharper results and a safer procedure (Sinigaglia et al., 2018).
In situ hybridization (ISH) methods rely on the application of an exogenous, conveniently labeled, nuclei acid fragment (called a probe) to fixed tissues or cells. A combination of high temperatures and solvents will promote the separation of native nuclei acid strands (a process called denaturation), and the annealing of probes to their endogenous targets (the hybridization). Only the paired probes can then be detected, thus revealing the localization of the target sequence. This versatile method allows, for example, the ability to identify which cells within a tissue are expressing a certain gene or to detect chromosomic abnormalities, and it has broad application in research, diagnostics, and teaching.
The first study presenting this method employed sodium hydroxide to denature the DNA of Xenopus oocytes and facilitate hybridization of radioactively-labeled probes (Pardue and Gall, 1969). Later works tested a variety of denaturing agents, but after a period of experimentation, the use of 50-70% formamide in the hybridization buffer became the standard. Formamide is an efficient but hazardous solvent which entails the application of strict safety procedures for manipulations and waste disposal.
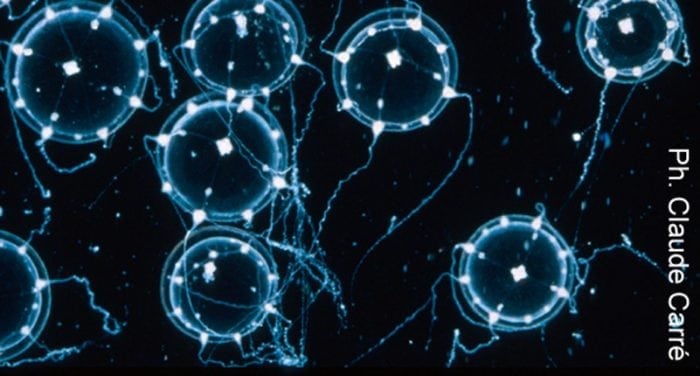
Our laboratories study developmental biology and the evolution of diverse marine species, including the hydrozoan jellyfish Clytia hemisphaerica (in France), the worm Priapulus caudatus, and the lamp shell Terebratalia transversa (in Bergen). Research on lesser-known animals can be very rewarding but also poses unexpected challenges. Jellyfishes are fragile organisms, and unfortunately, the standard ISH protocol, which had already been successfully employed to study gene expression in Clytia embryos and isolated organs, turned out to be rather unfit for the medusa, whose umbrella was highly damaged in the hybridization step. More worryingly, in many cases, we were unable to distinguish within our crumpled samples the real signal from the widespread background noise.
Months of frustrating troubleshooting pushed us to re-examine the essential steps of ISH and to investigate the research behind every reagent. The discovery that the hybridization buffer, now undisputed, had been different in the past prompted us to look more closely at its composition. We realized that another reagent could have denaturing properties similar to formamide – and was indeed employed as equivalent in other laboratory techniques: urea.
Urea is an organic compound, which is naturally produced by our livers or readily synthesized artificially, for applications in agriculture or industry. It is much safer than formamide and can be handled even by pregnant women (of course, basic safety rules still apply). This seemed like a promising candidate for a less aggressive treatment. We substituted the formamide in our original hybridization buffer with an equal volume of 8M urea solution, leaving the rest of the protocol unchanged.
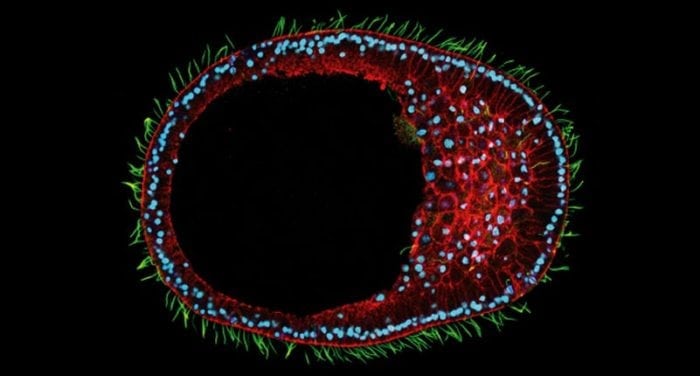
Clytia hemisphaerica early gastrula embryo. Credit: Tsuyoshi MOMOSE / Movincell
With excitement, we saw that the morphology of medusae was well preserved after the days-long exposure to the heated hybridization buffer. Upon signal detection, we obtained a beautiful and sharp staining, revealing previously unnoticed areas of expression for some genes. The background had almost entirely disappeared.
Given that such a simple change greatly improved our results, we wondered whether this alternative protocol could also help the study of other organisms. We tested the urea-based hybridization buffer on other marine animals (the worm P. caudatus, and two lamp shell species – T. transversa and Novocrania anomala), for which ISH protocols had already been established, but where aspecific staining was a common issue. In all cases, the signal-to-noise ratio improved, confirming previously detected patterns of gene expression and identifying areas of non-specific signal.
Numerous laboratories working on a variety of research organisms are today adopting the urea-based in situ hybridization protocol. The minimized hazard exposure and the reduced production of toxic waste, in fact, prompted also researchers who had no previous experimental issues to test this alternative. The same reasons induced the university teaching staff at the Institut de la Mer de Villefranche-sur-mer (IMEV, France) to adopt the new buffer in their practical courses.
Often people question the importance of studying animals other than mice or fish. These results show how research on less well-known organism provides us with new eyes for looking at the world and can have greater repercussion that goes beyond a restricted research field.
These findings are described in the article entitled A safer, urea-based in situ hybridization method improves detection of gene expression in diverse animal species, recently published in the journal Developmental Biology. This work was conducted by Chiara Sinigaglia, Evelyn Houliston, and Lucas Leclère from the Laboratoire de Biologie du Développement de Villefranche-sur-mer, and Daniel Thiel and Andreas Hejnol from the University of Bergen.
References:
- Sinigaglia C., Thiel C., Hejnol A., Houliston E., Leclère L., A safer, urea-based in situ hybridization method improves detection of gene expression in diverse animal species. Developmental Biology. 2018; 434(1):15-23. https://linkinghub.elsevier.com/retrieve/pii/S0012160617303755
- Pardue ML, Gall JG., Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proceedings of the National Academy of Sciences of the United States of America. 1969; 64(2):600-604.








