
Eukaryotic proteins are made up of both “ordered” and “disordered” parts. The former have a fixed 3D structure, while the latter do not take on a 3D structure during some of their functioning in cells and tissues. Such “intrinsically disordered regions” (IDRs) remain unfolded under physiological conditions, sampling lots of different diffuse shapes and conformations. IDRs have led to a profound reassessment of the protein structure-function paradigm, and their improper functioning can lead to many diseases. IDRs can mediate interaction between proteins in cells, including such diverse proteins as kinases, transcription factors, and translation inhibitors. They can also enable interaction with DNA and RNA.
Proteins are often chemically modified in cells immediately after translation. Such multifarious ‘post-translational’ modifications alter protein functioning and are placed at specific amino-acid residue sites. IDRs of proteins are more efficient homes for these modifications than fixed 3D structures are since more of the modifications can be arrayed in a stretch of protein sequence in an accessible way. Post-translational modifications can help to induce disorder-to-order transitions, that is, at moments when IDRs become ordered upon binding to a partner molecule. Furthermore, post-translational modifications in IDRs have key roles in cellular signaling and gene regulation. For example, complex patterns of post-translational modification on histone proteins modulate their interactions with DNA, affect nucleosome stability and thus regulate gene expression.
In our article [1], we studied three abundant and important types of modification (methylation, acetylation, and ubiquitination, abbreviated as “MAU”). These occur mainly at the positively-charged amino acids, arginine, and lysine. They are the three major modifications, next to phosphorylation and glycosylation, which regulate eukaryotic protein function. The interplay between modifications at different MAU sites facilitates complex regulatory programs involving both histone and non-histone proteins. However, the evolution of these MAU sites across eukaryotic species has not been well understood.
In [1], we exploited the recent explosion of new, rapidly-sequenced genome data for several hundred eukaryotes, to analyze the evolutionary appearance and conservation across the eukaryotic domain of amino-acid residues that are modified in this way in humans. Analysis of conservation across such a large panel of eukaryotes can give us more comprehensive insights into the evolutionary history of post-translational modifications while avoiding issues of data set completeness that may be a problem for experimental determination of MAU sites across a variety of complex multi-cellular species. We discovered that there is enough information in this new genome data to discern clear “signals” of conservation and emergence of these modifications over eukaryotic evolution. We analyzed the distribution of more than 15,000 MAU sites in ordered and disordered parts and probed their conservation across the hundreds of millions of years of eukaryote evolution (Figure 1).
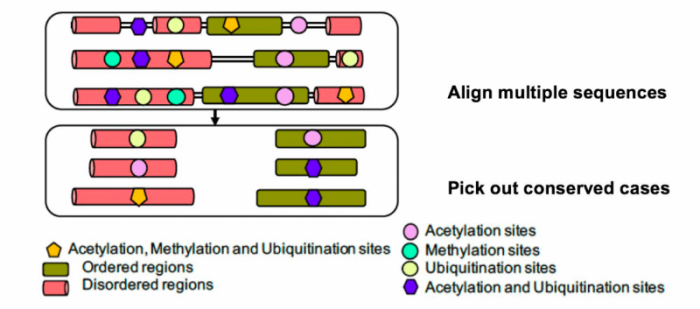
Figure 1: Proteins from several hundred eukaryotes are aligned on top of each other, then conserved modification sites are picked out of the alignments. Image courtsey Paul M. Harrison
Conservation signals were discovered that show us the maintenance and brand new emergence of MAU sites, across the whole evolutionary timescale of eukaryotes down to the appearance of the ape superfamily. We discovered that MAU sites are a major driving force for conservation of arginines and lysines in both the ordered and disordered parts of proteins. Surprisingly, we found that acetylation and ubiquitination cause conservation of lysine residues in IDRs, much more so than in ordered protein parts; this is despite IDRs generally being much less conserved than the ordered parts generally. Notably, we found evidence for the emergence of new modification sites on a large scale at different time points in eukaryotic evolution. For example, lysine modification sites appeared in the disordered parts of proteins after the dawn of deuterostomes, some 550 million years ago. For histones specifically, MAU sites have an idiosyncratic conservation pattern that is obvious since the emergence of mammals, that we can correlate with specific evolutionary time points.
These findings and more are described in the article entitled Discerning evolutionary trends in post-translational modification and the effect of intrinsic disorder: Analysis of methylation, acetylation and ubiquitination sites in human proteins, recently published in the journal PLOS Computational Biology. This work was conducted by Mohanalakshmi Narasumani and Paul M. Harrison from McGill University.
Reference:
- Narasumani & Harrison, “Discerning evolutionary trends in post-translational modification and the effect of intrinsic disorder: Analysis of methylation, acetylation and ubiquitination sites in human proteins”, PLOS Comp. Biol., https://doi.org/10.1371/journal.pcbi.1006349.









