
The zika virus (ZIKV) has come to recent attention because of unusual and severe neurological pathogenicity observed in recent epidemics in Asia and the Americas1,2 and in particular the discovery of congenital brain malformation in fetuses from mothers infected with ZIKV in the first 3 months of pregnancy3.
The absence of approved antiviral treatments or vaccines to treat or prevent ZIKV infection makes a compelling case to develop new strategies to combat this emerging health threat [eg. see 4].
Zika virus is closely related to Dengue and West Nile viruses
As a typical flavivirus, ZIKV is most closely related to dengue virus (DENV) and West Nile Virus (WNV), with the highest conservation of sequence in non-structural protein 5 (NS5)5, the methytransferase (MTase)/RNA-dependent RNA polymerase (RdRp) essential for viral replication in the cytoplasm6-8. NS5 from many flaviviruses, including DENV serotypes 1-4, Japanese Encephalitis Virus (JEV), and WNV [eg. 9,14], is known to shuttle between the nucleus and cytoplasm, with roles in the nucleus that include suppressing the antiviral response9-12, at least in part through effects on mRNA splicing of antiviral response genes [eg. 13]. In the case of DENV NS5, nuclear entry occurs through a well-characterized pathway dependent on the host importin (IMP) α/β heterodimer, preventing NS5 nuclear import through mutation, or agents that inhibit the activity of IMPα results in drastically reduced DENV virus production in infected cells9,12,14.
Significantly, the key basic amino acid residues of the central “nuclear localisation signal” within NS5 responsible for nuclear targeting in the case of DENV2 are highly conserved in NS5 from all DENV serotypes, but also, importantly, in related flaviviruses, including clinically relevant Asia and African ZIKV strains (see Fig. 1A). This raises the exciting possibility that anti-DENV agents targeting NS5 nuclear import could also impact ZIKV.
4-HPR inhibits ZIKV NS5 recognition by IMPs
We previously identified N-(4-hydroxyphenyl) retinamide (4-HPR) in a high-throughput screen for agents able to inhibit DENV NS5 recognition by IMPα/β, and showed it to be a potent inhibitor of infection with all DENV, including severe (haemorrhagic) forms of disease in an animal model as well as human disease-relevant model (peripheral blood mononuclear cells)14,16,17, underlining the key importance of this interaction for viral infection. 4-HPR also limits WNV infection17. Importantly, 4-HPR has an establish safety profile17-19 through the fact that, unlike agents targeting IMPs, it targets DENV NS5, and hence should not be toxic to mammalian cells. We reasoned that, based on the homology of NS5 from DENV and ZIKV (Fig. 1B, C), ZIKV NS5 should be able to be recognized with high affinity by IMPα/β in a similar fashion to DENV NS5, and that this should be able to be inhibited by 4-HPR. Experiments testing this directly confirmed that ZIKV was very similar to DENV NS5 in interacting with the IMPα/β heterodimer with high affinity (Fig. 2A; Table 1), and showing potent inhibition of this interaction by 4-HPR (Fig. 2B; Table 1).
4-HPR is a potent anti-ZIKV agent
Based on the observations above, we tested the ability of 4-HPR to inhibit ZIKV infection. Results for estimation of viral genomes or infectious virus showed that 4-HPR is a potent inhibitor of ZIKV infection (EC50 of c. 3 µM, Fig. 3). The fact that this EC50 value is close to the IC50 value for inhibition of ZIKV NS5-IMPα/β binding suggests that the mechanism of action of 4-HPR is through blocking ZIKV NS5 nuclear accumulation. The results imply that ZIKV NS5 nuclear targeting through IMPα/β is likely critical to infection17, presumably through its action on infected host cell transcription and thereby the antiviral response, in a similar fashion to DENV NS59-13. Excitingly, 4-HPR was recently reported to protect against ZIKV in an animal model20.
Future directions
4-HPR is clearly a therapeutic agent with exciting potential not only to treat DENV infection, but also ZIKV infection, and other flavivirus infections. 4-HPR’s safety record in humans17-19 makes the progression of 4-HPR to the clinic for DENV and potentially ZIKV a high priority for the future.
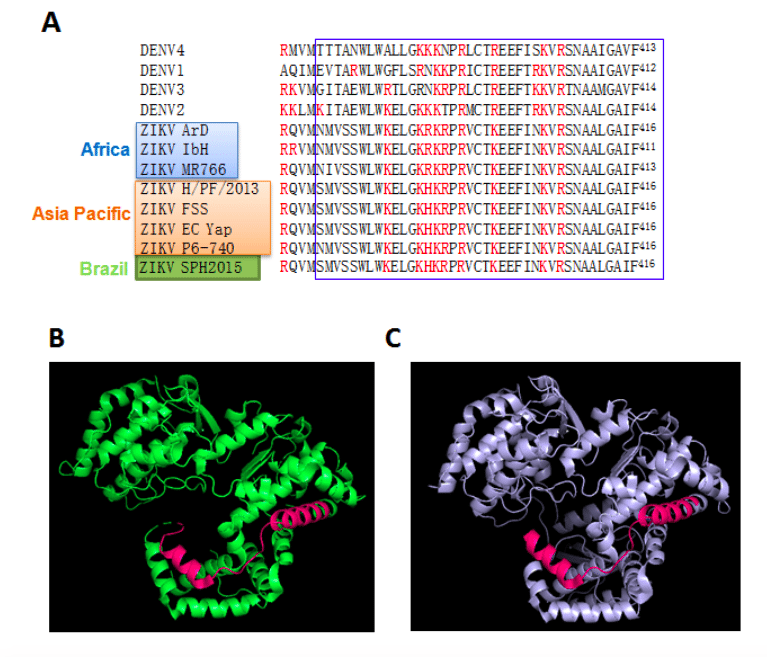
Figure 1. Close homology of key regions of ZIKV NS5 to DENV NS5. A. Alignment of the key domain for nuclear trafficking of DENV NS5 (the “nuclear localization signal” or NLS 17-14) to the homologous regions from ZIKV strains. The single letter amino acid code is used, with the NLS boxed in blue and key basic residues highlighted in red. Sequences are: DENV1-4 (GenBank IDs AIU47321.1, AAK67712.1, AHN50411.1, AMP43484.1 respectively), and ZIKV ArD_41519 (ZIKV ArD; Senegal, 1984; GenBank: AEN75266.1), ZIKV IbH-30656 (ZIKV IbH; Nigeria, 1968; GenBank: AMR68906.1), ZIKV MR766 (Africa; GenBank: AY632535.2), ZIKV H/PF/2013 (French Polynesia, 2013; GenBank: KJ776791.2), ZIKV FSS13025 (ZIKV FSS; Cambodia, 2010; GenBank: AFD30972.1), ZIKV EC Yap (Micronesia, 2007; GenBank: EU545988.1), ZIKV P6-740 (Malaysia, 1966; GenBank: ANK57896.1) and ZIKV SPH2015 (Brazil; GenBank:KU321639.1). It should be noted that the “NLS-like” sequence within the C-terminus of DENV2 NS521 is not conserved in ZIKV NS5 or indeed the majority of DENV or WNV etc. NS5 sequences. B, C. Models from X-ray crystallographic structures of ZIKV NS5RdRp MR766 (Africa; PDB: 5U04; B) and DENV3 NS5RdRp (PDB: 4HHJ; C) represented in cartoon diagram with basic NLS region highlighted in pink. This figure was generated using PyMol [http://www.pymol.org].
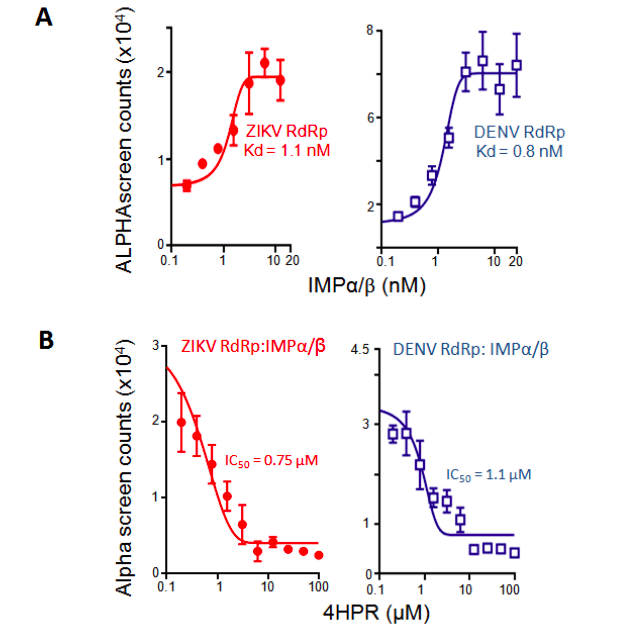
Figure 2. IMPs bind ZIKV NS5 directly with high affinity; inhibition by 4-HPR. ALPHAscreen technology was used to estimate the dissociation constant (Kd) of binding of IMPα/β to His6-ZIKV SPH2015 strain (Brazil) NS5 protein amino acids 273-887 (RdRp) or His6-DENV RdRp12 (A), and EC50 for inhibition by 4-HPR (B) as indicated. Data represent the mean ± SD for triplicate wells from a single typical experiment (see Table 1 for pooled data).
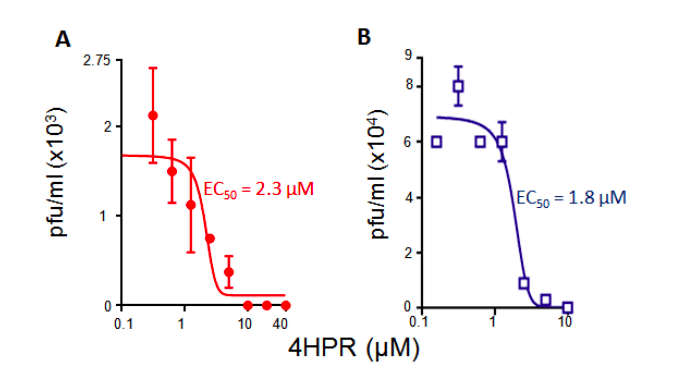
Figure 3. 4-HPR inhibits ZIKV infection. Cells were infected with ZIKV (Asian/Cook Islands/2014) or DENV2 together with increasing concentrations of 4-HPR, and the infectious virus titres determined by plaque assay12. Results represent the mean ± SD (n = 2), representative of >2 independent experiments.
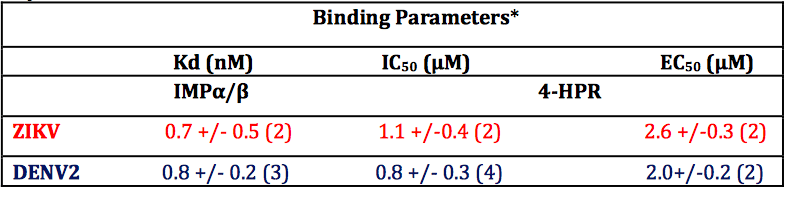
Table 1. Summary of binding, IC50 and EC50 data for 4-HPR from ALPHAscreen and plaque assay analysis. *Results represent the mean + SD (n) from the analysis as per Figure 2 (Kd, IC50) and 3 (EC50).
These findings are described in the article entitled Nuclear import inhibitor N-(4-hydroxyphenyl) retinamide targets Zika virus (ZIKV) nonstructural protein 5 to inhibit ZIKV infection, recently published in the journal Biochimica Biophysica Research Communications.
This work was conducted by Chunxiao Wang, Sundy N.Y. Yang, and David A. Jans from Monash University, and Kate Smith and Jade K. Forwood from Charles Sturt University.
References:
- G.S. Campos, A.C. Bandeira, S.I. Sardi, Zika Virus Outbreak, Bahia, Brazil. Emerg. Infect. Dis. 21 (2015) 1885-1886.
- M. Hennessey, M. Fischer, J.E. Staples, Zika Virus Spreads to New Areas – Region of the Americas, May 2015-January 2016, MMWR Morb Mortal Weekly Rep. 65 (2016) 55-58.
- J. Mlakar, M. Korva, N. Tul, M. Popovic, M. Poljsak-Prijatelj, J. Mraz, M. Kolenc, K.R. Rus, T.V. Vipotnik, V.F. Vodušek, A. Vizjak, J. Pižem, M. Petrovec, T.A. Županc, Zika Virus Associated with Microcephaly, New England J. Med. 374 (2016) 951-958.
- V. Boldescu, M.A.M. Behnam, N. Vasilakis, C.D. Klein, Broad-spectrum agents for flaviviral infections: dengue, Zika and beyond, Nature Rev. Drug Disc. 16 (2017) 565-586.
- Z. Zhu, J.FW. Chan, K-M. Tee, G.K-Y. Choi, S.K-P. Lau, P.C-Y Woo, H. Tse, K-Y. Yuen, Comparative genomic analysis of pre-epidemic and epidemic Zika virus strains for virological factors potentially associated with the rapidly expanding epidemic, Emerging Microbes Infect. 5 (2016) e22.
- B. Wang, X.F. Tan, S. Thurmond, Z.M. Zhang, A. Lin A, R. Hai, J. Song, The structure of Zika virus NS5 reveals a conserved domain conformation, Nature Comm. 8 (2017) 14763.
- A.S. Godoy, G.M. Lima, K.I. Oliveira, N.U. Torres, F.V. Maluf, R.V. Guido, G. Olivia, Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase, Nature Comm. 8 (2017) 14764.
- AK. Upadhyay, M. Cyr, K. Longenecker, R. Tripathi, C. Sun, D.J. Kempf, Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5, Acta Cryst. Section F, Struct. Biol. Comm. 2017; 73 (2017) 116-122.
- M.J. Pryor, S.M. Rawlinson, R.E. Butcher, C.L. Barton, T.A. Waterhouse, S.G. Vasudevan, P.J. Wright, D.A. Jans, Nuclear localization of dengue virus nonstructural protein 5 through its importin a/b-recognized nuclear localization sequences is integral to viral infection, Traffic 8 (2007) 795-807.
- S.M. Rawlinson, M.J. Pryor, P.J. Wright, D.A. Jans, CRM1-mediated nuclear export of dengue virus NS5 RNA polymerase regulates the kinetics of virus production, J. Biol. Chem. 284 (2009) 15589-15603.
- S.M. Best, The Many Faces of the Flavivirus NS5 Protein in Antagonism of Type I Interferon Signaling, J Virol. 91 (2017) e01970-16.
- J.E. Fraser, C. Wang, K.K.W. Chan, S.G. Vasudevan, D.A. Jans, Novel Dengue Virus Inhibitor 4-HPR Activates ATF4 Independent of Protein Kinase R–like Endoplasmic Reticulum Kinase and Elevates Levels of eIF2α Phosphorylation in Virus Infected Cells, Antiviral Res. 130 (2016) 1-6.
- F.A. De Maio G. Risso, N.G. Iglesias, P. Shah, B. Pozzi, L.G. Gebhard, P. Mammi, E. Mancini, M.J. Yanovsky, R. Andino, N. Krogan, A. Srebow, A.V. Gamarnik, The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing, PLoS Pathog. 12 (2016) e1005841.
- M.Y.F. Tay, J.E. Fraser, W.K. Chan, N.J. Moreland, A.P. Rathore, C. Wang, S.G. Vasudevan, D.A. Jans, Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Res. 99 (2013) 301-306.
- J.K. Forwood, A. Brooks, L.J. Briggs, C-Y. Xiao, D.A. Jans, S.G. Vasudevan, The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site, Biochem Biophys Res Commun. 257 (1999) 731-737.
- K.M. Wagstaff, H. Sivakumaran, S. Heaton, D. Harrich, D.A. Jans, Ivermectin is a specific inhibitor of importin a/b-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochem. J. 443 (2012) 851-856.
- J.E. Fraser, S. Watanabe, C. Wang, K.K. Chan, B. Maher, A. Lopez-Denman, C. Hick, K.M. Wagstaff, P. Sexton, J. Mackenzie, S.G. Vasudevan, D.A. Jans, A nuclear transport inhibitor that modulates the unfolded protein response and provides in vivo protection against lethal Dengue virus infection, J. Infect. Dis. 210 (2014) 1780-1791.
- B.J. Maurer, M.H. Kang, J.G. Villablanca, J. Janeba, S. Groshen, K.K. Matthay, P.M. Sondel, J.M. Maris, H.A. Jackson, F. Goodarzian, H. Shimada, Phase I trial of fenretinide delivered orally in a novel organized lipid complex in patients with relapsed/refractory neuroblastoma: a report from the New Approaches to Neuroblastoma Therapy (NANT) consortium, Ped. Blood Cancer 60, (2013) 1801-1808.
- J.G. Villablanca, W.B. London, A. Naranjo, P. McGrady, M.M. Ames, J.M. Reid, R.M. McGovern, S.A. Buhrow, H. Jackson, E. Stranzinger, B.J. Kitchen, Phase II study of oral capsular 4-hydroxyphenylretinamide (4-HPR/fenretinide) in pediatric patients with refractory or recurrent neuroblastoma: a report from the Children’s Oncology Group, Clin. Cancer Res.17 (2011) 6858-6866.
- J.D. Pitts, PC Li, M. de Wispelaere, P.L. Yang. Antiviral activity of N-(4-hydroxyphenyl) retinamide (4-HPR) against Zika virus. Antiviral Res. 147(2017) 124-130.
- M.Y.F. Tay, K. Smith, I.H.W. Ng, K.W.K. Chan, Y. Zhao, E.E. Ooi, J. Lescar, D. Luo, D.A. Jans, J.K. Forwood, S.G. Vasudevan, The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production, PLOS Path. 12 (2016), e1005886.









