
Accordingly to the World Health Organization (WHO), almost 2 billion adults were overweight in 2016. Of these, over 650 million were obese. Clearly, overweight and obesity have become a global emergency with serious health concerns for affected individuals. Indeed, being overweight or obese increases the risks of diabetes, hypertension, atherosclerosis, and cancer. Considering the health consequences of excess fat accumulation, there is an urgent need to develop new strategies to counteract weight gain.
Obesity develops when energy intake exceeds energy expenditure, leading to increased body mass. For this reason, current therapeutic strategies aim at restricting energy uptake and absorption, thus generating a negative energy balance with a consequent decrease in body mass. However, so far, these approaches have shown limited efficacy, and alternative strategies are urgently needed to increase energy dissipation.
Genetic heritability only partially explains the growing number of obese individuals, and much attention has been given to environmental factors and epigenetic changes. Epigenetics are heritable changes in gene expression that occur without alteration in DNA sequence and exhibit plasticity to the environment. It is well documented that epigenetic variants affect individual responses to environmental stimuli and the risk to develop obesity and other metabolic diseases.
Obesity occurs when fat accumulates within the body. We can distinguish two types of fat, with different metabolic activity. The white fat (WAT) mainly stores energy and is increased in obesity. On the contrary, the brown one (BAT) is able to burn excess of energy introduced with food. Adult humans have active BAT deposits, and very recently, it was shown that this tissue increases its glucose uptake after a meal. Indeed, activated human BAT improves whole-body glucose homeostasis and insulin sensitivity and can lead to reduced body weight, indicating that it plays an important role in energy balance. Consequently, a better understanding of the molecular control of BAT function may allow one to address obesity and metabolic disorders.
BAT activity is regulated by the peroxisome proliferator-activated receptor gamma (PPAR-γ) transcription factor, which is highly expressed within the tissue and controls glycemic metabolism, adipogenesis, energy balance, and lipid biosynthesis. Importantly, PPAR-γ expression levels decrease in obese patients with diabetes. Although a pharmacological approach to increase PPAR-γ expression is available as a mean to increase insulin sensitivity, these drugs present several side effects, increasing the urgency to find alternative ways to regulate PPAR-γ in order to develop safer and more effective diabetes therapeutics.
In our work, we found that two histone methyltransferases, named Suv420h1 and Suv420h2, regulate metabolism and body weight. These two enzymes are responsible for chromatin compaction and consequent gene repression. Several studies previously reported a possible involvement of Suv420h proteins in metabolism regulation. Yet, a direct link between Suv420h activity and metabolism was still lacking.
We generated a mouse model in which Suv420h enzymes were specifically deleted in the adipose tissue. Knockout mice showed improvement of several metabolic parameters, such as increased BAT mitochondrial activity, better glucose homeostasis, and reduced adipose tissue, suggesting an increased metabolic activity. Indeed, we found that mice lacking the two enzymes were resistant to diet-induced obesity. When fed with a fat-rich diet, the mutant mice did not gain weight at the same extent as the control mice, in which Suv420h enzymes were normally expressed.
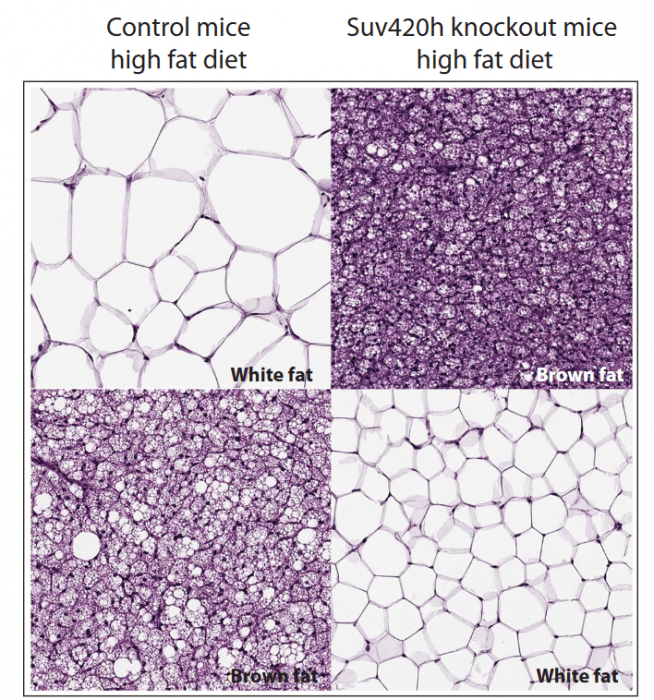
Figure courtesy Simona Pedrotti
Bioinformatic analysis of gene expression changes in BAT isolated from control and knockout mice after high-fat diet feeding revealed upregulation of genes involved in metabolism control that confers protection toward diet-induced obesity. Moreover, the majority of the upregulated genes were indeed PPAR-γ targets, the master regulator of brown fat activity. We then hypothesized that PPAR-γ expression could be regulated by Suv420h. We found upregulation of PPAR-γ concomitant to Suv420h downregulation, and chromatin immunoprecipitation assay (ChIP) showed that Suv420h directly bind to PPAR-γ transcription start site inhibiting its expression.
These results support the hypothesis that PPAR-γ is a direct target of Suv420h. Indeed, by using a specific inhibitor of Suv420h activity in cells directly isolated from brown fat, we found that PPAR-γ activity was increased, resulting in augmented mitochondrial activity and energy expenditure. Our findings clearly demonstrated that Suv420h enzymes are candidate targets for obesity and weight management, opening new strategies for future therapeutics.
These findings are described in the article entitled The Suv420h histone methyltransferases regulate PPAR-γ and energy expenditure in response to environmental stimuli, recently published in the journal Science Advances.









