
In the light of numerous recent reports of rapid extinction of animal and plant species on our planet, it is critical to understand the dynamics of speciation — that is, how species are formed and maintained.
Many scenarios of speciation have been proposed and analyzed. For example, speciation can be facilitated by physical/geographic isolation of the diverging populations, by adaptation to different environments, by the formation of unfit hybrids, or by the evolution of assortative mating (individuals tend to reproduce with individuals that are similar to themselves).
One intriguing and contentious scenario in which new species can arise is termed hybrid speciation. In this method, following the contact of two species, the hybrid population that is generated becomes genetically different from both parental sources. This process results in a third, independent, new species. Prominent examples of potential hybrid species include the Heliconius butterfly, the Italian sparrow, and the Helianthus sunflower. However, the conditions in which hybrid speciation can occur are likely restrictive. This is illustrated by a proposed theoretical model that explains how hybrid speciation can occur. Importantly, the model requires the two parental populations to be different enough that they are considered separate species, but at the same time compatible enough that they can form the necessary hybrid individuals.
Going more into detail, we wanted to understand under which specific conditions on the genomes of the hybridizing parental species hybrid speciation is most likely to happen. To simplify the problem, we built a minimal mathematical model of hybrid speciation. A minimal model reproduces a biological phenomenon observable in nature with the smallest number of ingredients possible. Building and analyzing a minimal model allows a researcher to capture and understand the key elements of a biological phenomenon.
In our work, we focused on a specific form of reproductive isolation, namely postzygotic isolation. When there is postzygotic isolation, individuals from two species can mate with each other but the resulting offspring are either not viable or infertile. A famous example of postzygotic isolation between species is the mule, which results from the cross between a horse and a donkey. Mules are infertile and, therefore, represent an evolutionary dead-end (any gene carried by a mule will not be transmitted to any offspring).
The most well-supported model of how postzygotic isolation can evolve is the Bateson-Dobzhansky-Muller model (named after its inventors). This model describes how random genetic changes that occur in geographically isolated populations can display detrimental effects only when brought together in a hybrid. In a compelling way, the Bateson-Dobzhansky-Muller model explains how natural selection cannot “see,” and thus act against the resulting postzygotic genetic incompatibilities (as we term those interacting genetic changes) until they are exposed in hybrid individuals.
For reproductive isolation with respect to the two parental species, and thus hybrid speciation, we need at least two pairs of such genetic incompatibilities; one to isolate the new species from each of its parental populations. Therefore, we model the evolution of four diallelic loci. A diallelic locus is a position in the genome with two possible variants. At each of these loci, we label as “ancestral” the genetic variant that was present in the ancestral population and “derived” the variant that appeared after mutation. Postzygotic isolation results from genetic incompatibilities between the derived variants that are specific to each parental populations. To study the best conditions for hybrid speciation, we used computer simulations.
We showed that relative location of the genetic incompatibilities in the genome is critical to the probability of observing hybrid speciation. Specifically, the incompatibilities need to be located apart from each other, but not too far apart. In addition, the order of the four involved loci in the genome can crucially change the hybrid speciation probability, as illustrated in Figure 1.
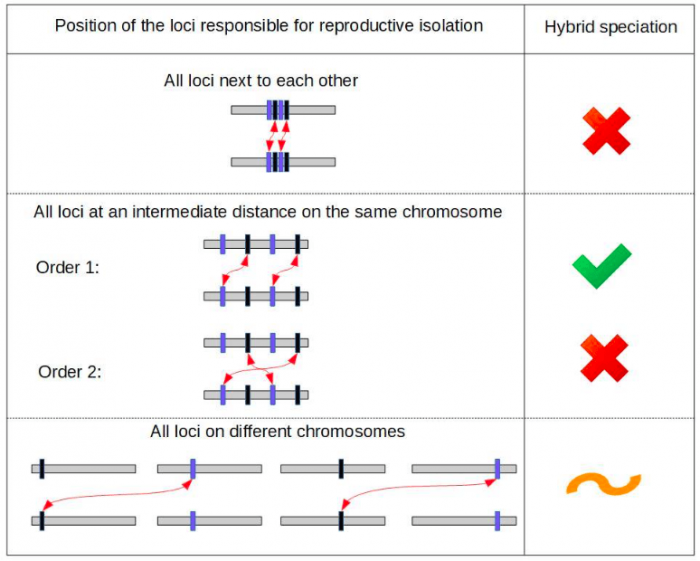
Figure 1: Hybrid speciation becomes very likely only for specific orders and positioning of the loci responsible for reproductive isolation along the genome. Here, the green checkmark indicates a scenario in which hybrid speciation is highly likely, the orange tilde that is unlikely and the red crosses that is extremely unlikely. It is surprising that only a slight change in the ordering of the loci on a chromosome can change the hybrid speciation probability dramatically; this result was revealed through building a minimal model and the use of computer simulations. The red arrows indicate the interacting loci of the two genetic incompatibilities. We represented here only two of the six possible orderings of the four loci. Image credit: Alexandre Blanckaert
These results can explain the contradictory observations from the empirical literature: on the one hand, hybrid speciation is rarely observed; on the other hand, multiple hybrid speciation events have been reported between the same species. For example, three independent instances of hybrid speciation have been reported for the Italian sparrow. Similarly, hybrid speciation has been observed between two species of yeast both in nature and in the laboratory. Consistent with our predictions, these findings highlight the occasional repeatability of the hybrid speciation process. This indicates that the occurrence of hybrid speciation is not simply random: the genetics of some species may make some species pairs more prone to founding a new hybrid species than others.
Our computational work provides a potential explanation for this pattern, which may help to settle the ongoing debate in the research community about the probability of hybrid speciation. Furthermore, our work provides hypotheses about the best conditions for hybrid speciation, which can in the future be tested empirically.
With respect to conservation biology, hybridization is an important issue because it can both harm and aid the maintenance of biodiversity. Many species have been introduced in a new habitat due to human activities. These invasive species can compete or hybridize with the native ones, with a risk of losing the local species. Extrapolating our results to this question, some species should be able to maintain their genetic integrity and will therefore mainly compete for resources, whereas others are permissive to the integration of genetic material from the invasive species. Which of these scenarios applies, and whether this process is beneficial or detrimental is likely to depend critically on the “genetic architecture,” i.e., the location of interacting and potentially incompatible loci in the genome.
Our work, therefore, implies that knowing how two species are reproductively isolated from each other may be crucial to predicting the outcome of hybridization. This, in turn, is key to designing appropriate conservation strategies under the threat of invasive species.
These findings are described in the article entitled In search of the Goldilocks zone for hybrid speciation, recently published in the journal PLOS Genetics. This work was conducted by Alexandre Blanckaert and Claudia Bank from the Instituto Gulbenkian de Ciência.









