
Malignant melanoma is a skin cancer primarily caused by overexposure to UV rays from the sun or tanning beds. Unfortunately, the number of skin cancers worldwide has increased dramatically over the past decades. The main factors for this trend are simply poor practice when protecting the skin from the sun, the extensive use of tanning beds, skin type, and the increase of people traveling abroad to spend their holidays in the sun.
Due to its tendency to spread rapidly and its resistance to drugs, malignant melanomas rank among the deadliest cancers. From a molecular point of view, malignant melanoma are primarily caused by mutations of components that reside in the so-called MAPK signalling pathway. More than half of malignant melanomas are caused by activating mutations in the BRAF gene (such as BRAFV600E), while around one quarter is caused by mutations in the NRAS gene.
Targeted treatments in form of small molecule drugs (e.g. Dabrafenib, Vemurafenib) exist for BRAF-driven melanomas, often in combination with MEK inhibitors (Trametinib). However, no direct targeted therapies are currently in place or approved to treat melanomas caused by mutations in the NRAS gene, leaving these melanomas extremely difficult to treat. Current clinically used RAF inhibitors are ineffective in RAS-mutant tumours because they enhance homo- and heterodimerization of RAF kinases, leading to paradoxical activation of oncogenic MAPK signalling. In a RAF dimer, one protomer is drug bound and allosterically activates the other, drug-free protomer conferring resistance. Although the emergence of different drug affinities between monomers and protomers in a dimer is enigmatic, it can be explained by thermodynamics (Kholodenko, 2015). Overcoming enhanced RAF dimerization and the resulting resistance is a challenge for drug design.
An international team based at the institute of Systems Biology Ireland (University College Dublin, Ireland) together with international collaborators in the UK, US, and Greece, now used a combined experimental and computational approach to solve this important puzzle.
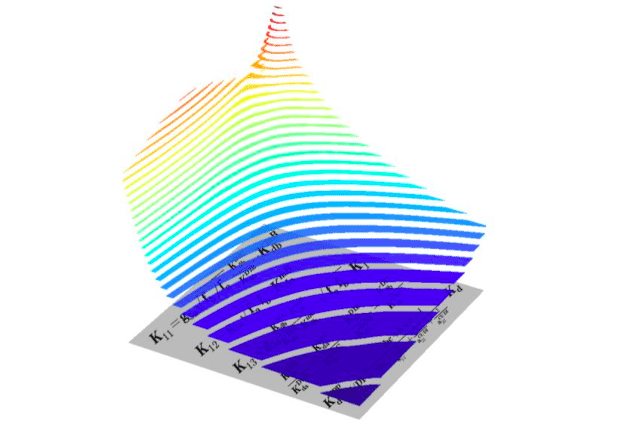
The figure shows oncogenic signalling responses (ppERK, vertical z-axis) to two different RAF inhibitors (horizontal x- and y-axes) that synergise in inhibition of MAPK pathway in RAS-mutant cell. Each drug alone induces paradoxical activation and inhibits MAPK pathway only in very high doses, but together these drugs effectively inhibit it. Concave isolines on the figure are called Loewe isoboles and represent synergy between drugs. Below you can see some key formulas of the mathematical model presented in the paper. (Credit: Oleksii Rukhlenko)
For this, they set out to generate a next-generation computer model of the underlying molecular signalling network which not only integrates the known protein-protein interaction and network architecture, but also includes the thermodynamics and kinetics of drug- protein interactions, spontaneous intra-molecular motions of proteins, structural elements of the protein targets, post-translational modifications of the signalling molecules involved, and the cell mutational status. In this so-called rule-based modelling approach, a staggering 25,000 reactions were generated. This computer model was then used to simulate and predict suitable combinations of distinct RAF inhibitors with the aim to target malignant melanoma more efficiently.
Computer predictions were then successfully validated in cell line models representing different malignant melanoma subclasses. Their results suggest which combinations of RAF inhibitors are suitable for particular genetic backgrounds, and, importantly, demonstrate that specific RAF inhibitor combinations can be applied to overcome oncogenic RAS signalling. Model predictions go beyond malignant melanomas to all cancers bearing RAS-mutations (e.g. colorectal cancer, pancreatic cancer, non-small cell lung cancer, etc.). This so-called systems biology approach to drug design will hopefully streamline drug development in the future.
Clearly, these results do not mean doctors will start treating malignant melanoma or any other RAS-mutant cancer with these drugs combinations tomorrow, but it’s a very promising step in the right direction. As usual, there is still a lot of work to translate these results from discovery to treatment.
Research team:
These findings are described in the article entitled Dissecting RAF Inhibitor Resistance by Structure-based Modeling Reveals Ways to Overcome Oncogenic RAS Signaling, recently published in the journal Cell Systems. This large collaborative study was led by a number of research groups at Systems Biology Ireland including Prof. Boris Kholodenko (Dr. Oleksii S. Rukhlenko, Dr. Fahimeh Khorsand), Prof. Walter Kolch (Dr. Aleksandar Krstic, Dr. Nora Rauch), Dr. Jens Rauch (Dr. Cheree Fitzgibbon), and Dr. David Matallanas. International collaborators include Dr. Keesha Erickson and Prof. William Hlavacek at Los Alamos National Laboratory, Prof. Richard Posner at Northern Arizona University, Dr. Silvia Gómez-Coca and Prof. Edina Rosta at King’s College London, as well as Dr. Jan Rozanc and Prof. Leonidas Alexopoulos at ProtATonce Ltd (Athens, Greece).
Publication reference:
- O. S. Rukhlenko, F. Khorsand, A. Krstic, J. Rozanc, L. G. Alexopoulos, N. Rauch, K. E. Erickson, W. S. Hlavacek, R. G. Posner, S. Gómez-Coca, E. Rosta, C. Fitzgibbon, D. Matallanas, J. Rauch, W. Kolch, and B. N. Kholodenko, “Dissecting RAF Inhibitor Resistance by Structure-based Modeling Reveals Ways to Overcome Oncogenic RAS Signaling,” Cell Systems, Volume 7, Issue 2, 22 August 2018, Pages 161-179.e14, DOI: 10.1016/j.cels.2018.06.002.
You can read the full paper here:
https://www.cell.com/cell-systems/fulltext/S2405-4712(18)30242-4
Video:
A brief video describing this study can be found here:









