
Measuring the similarity between two graphs is a basic operation with many applications. For example, in the fields of chemical informatics and drug design, we often need to compare the similarity of a chemical (as modeled by a labeled graph) to a known drug (as modeled by another labeled graph) in order to evaluate the potential of the chemical for novel drug design. Applications include bioinformatics, data mining, pattern recognition, and graph classification.
Graph edit distance (GED) is a useful metric that gives a measure of similarity of two graphs. Intuitively, graph edit distance measures how many operations must be applied in order to transform the first graph into the second. Typical operations include inserting or deleting a node, inserting or deleting an edge, changing a node label, and changing an edge label. Computing the graph edit distance is an NP-hard problem, and thus computational cost is a concern, especially when we need to compute graph edit distances many times, as in some applications.
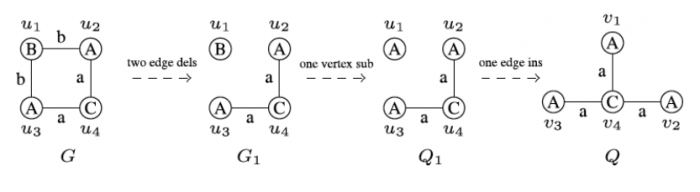
Figure 1: An Illustration of Graph Edit Distance. Figure republished with permission from Elsevier from doi.org/10.1016/j.knosys.2018.10.002
Several approaches have been developed for the efficient computation of graph edit distance. Typically, the possible graph mappings (between the two graphs) are organized as an ordered search tree, where the inner nodes denote partial mappings and the leaf nodes denote complete mappings. Riesen et al. recently proposed the A⋆-GED method. The method is vertex-based because the inner nodes (i.e., partial mappings) are extended in an iterative manner by extending unmapped vertices of the two graphs. The authors developed a heuristic function to estimate a lower bound on the GED in order to prune the search and reduce the number of mappings considered. Nevertheless, a major drawback of A⋆-GED is that it often needs to store too many partial mappings, resulting in huge memory consumption; in practice, A⋆-GED is only applicable to relatively small graphs.
In this paper, Chen and co-authors present BSS_GED, a novel vertex-based mapping method that calculates the GED in a reduced search space, which is enabled by more aggressively identifying and discarding invalid and redundant mappings. BSS_GED employs the beam-stack search paradigm, a widely utilized search algorithm in AI, combined with two specially designed heuristics to improve the GED computation, achieving a trade-off between memory utilization and expensive backtracking calls. Through extensive experiments, the authors demonstrate that BSS_GED is highly efficient on both sparse and dense graphs and outperforms the state-of-the-art methods. BSS_GED was further evaluated to solve the well-investigated graph similarity search problem. The experimental results show that this method is an order-of-magnitude faster than the state-of-the-art graph similarity search methods.
These findings are described in the article entitled An Efficient Algorithm for Graph Edit Distance Computation, recently published in the journal Knowledge-Based Systems, 163, January 2019, 762–775, doi.org/10.1016/j.knosys.2018.10.002. This work was conducted by Xiaoyang Chen, Hongwei Huo, Jun Huan, and Jeffrey Scott Vitter.








