
Science is often advanced with the development of new technologies. Since the sequencing of the first human genome, there has been much progress made in DNA sequencing technologies. We now have the ability to sequence complete genomes for a relatively low cost and much of the analyses can be done within a small research group. As a result, genomes are being sequenced across multiple taxonomic groups and research in genetics is quickly moving to a genomic scale. Studies that were once done with a few genetic markers are now using data from complete genomes, an approach which expands the scope of scientific questions that can be addressed.
To highlight the current state of genome sequencing within the Arthropoda, the journal Current Opinion in Insect Science published an issue dedicated to reviews of selected insect taxa. This series of articles focused on available genome sequences and future work necessary to accelerate the use of genomic technologies in entomological research. One particular review covered research within the Lepidoptera, the insect order comprised of butterflies and moths. This article not only reviews the current state of Lepidoptera genome sequencing but also emphasizes future challenges, including suggestions for storing and distributing genomic data to the arthropod research community.
The Lepidoptera (butterflies and moths) is one of the most ecologically diverse insect orders with more than 157,000 species described in 43 superfamilies. Most Lepidoptera belong to the taxonomic grouping of Ditrysia, which contains approximately 98% of the described species (Figure 1). Their genomes are relatively small in size (~200 – 800MB or 1⁄4 of the human genome) and lack structural complexity. However, there are < 80 species among <10 superfamilies whose genomes have been sequenced and assembled, with most belonging to butterfly, and a few moth families (Figure 1).
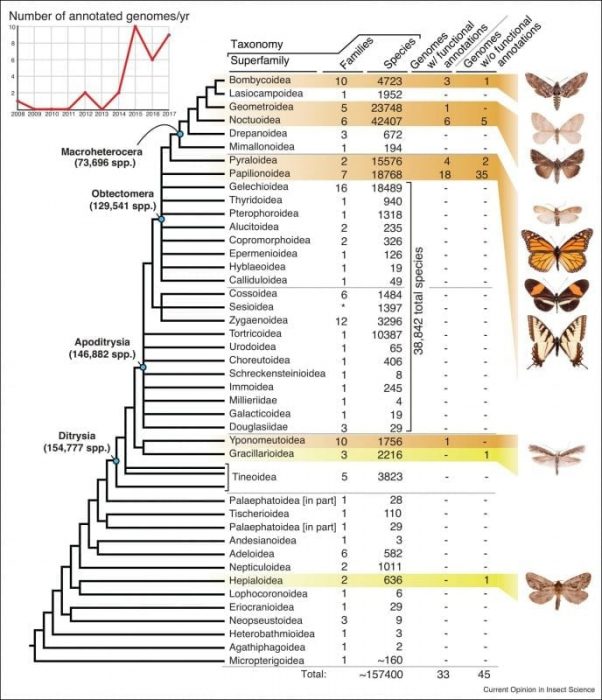
Figure 1. Phylogeny of Lepidoptera showing relationships among the major superfamilies and the number of assembled genomes (modified from Mitter et al. [4]). Orange highlights indicate superfamilies with at least one genome with a functional gene annotation; yellow indicates superfamilies with only a single genome and no functional annotation. The graph on the upper left shows the number of annotated genomes published per year since 2008. Republished with permission from Elsevier from https://doi.org/10.1016/j.cois.2017.12.004.
One central repository for Lepidoptera genomes is lepbase, which provides associated assembly statistics and gene annotations [1]. Platforms such as the i5k Workspace@NAL [2] hosts arthropod genomes and provide analytical assistance for users with limited bioinformatic experience. There are a number of other valuable databases available and often users need to search multiple sources to find a genome assembly of interest. Further complications include sequencing projects occurring in parallel without researchers being aware of related work. To avoid potential conflicts, it is recommended that members of lepbase or i5k be informed of genome sequencing projects to keep the community updated.
As to long-term data storage, it is good practice to archive completed or draft genome assemblies within the National Center for Biotechnology Information site (NCBI) upon completion to ensure that the data are screened and assigned an accession number for reference. It can be difficult to determine when a genome is “complete” and several versions of a single species’ genome can be released at different draft stages, which often makes comparisons difficult. With an assigned accession number, if improvements are made to a released genome, users can archive different versions of the same genome sequence and ensure downstream analyses are completed on a standardized set of genomic data.
The first Lepidoptera genome sequenced was the domesticated silkworm, Bombyx mori [3], a model species important for commercial silk production. Since then, the majority of Lepidoptera genomes have been sequenced within the past 5 years (Figure 1) and continues to grow as sequencing costs decrease and sequencing technologies improve. Broader sampling across major phylogenetic lineages is needed for the field of Lepidoptera genomics to move forward. Moreover, scientists should continue to make genomes publicly available along with metadata describing the assembly process while noting any limitations so they can be used more efficiently.
These findings are described in the article entitled Lepidoptera genomes: current knowledge, gaps and future directions, recently published in the journal Current Opinion In Insect Science. This work was conducted by Deborah A Triant, Scott D Cinel, and Akito Y Kawahara from the University of Florida in Gainesville, FL.
References:
- Challis RJ, Kumar S, Kumar K, Dasmahapatra K, Jiggins CD, Blaxter M. Lepbase: the Lepidopteran genome database. bioRxiv 2016 https://www.biorxiv.org/content/10.1101/056994v1
- Poelchau M, Childers C, Moore G, Tsavatapalli V, Evans J, Lee C-Y, Lin H, Lin J-W, Hackett K. The i5k Workspace@NAL—enabling genomic data access, visualization and curation of arthropod genomes. Nuc Acids Res 2015 43:D714-D719.
- Mita K, Kasahara M, Sasaki S, Nagayasu Y, Yamada T, Kanamori H. The Genome Sequence of Silkworm, Bombyx mori. DNA Res 2004 11:27-35.
- Mitter C, Davis DR, Cummings MP. Phylogeny and evolution of Lepidoptera. Annu Rev Entomol. 2017 62:265-283.









