Metabolomics aims at providing information about “metabolome,” the comprehensive small molecules of living organisms, which has been used for elucidating biochemical reactions, finding relationships between biological phenotypes, and linking metabolites to the upper omics hierarchies such as genomes, transcriptomes, and proteomes.
Recently, many studies have reported that metabolites themselves are deeply involved in physiological functions and homeostasis, including (1) oxylipins, an oxidized fatty acids group that acts as bioactive metabolites in inflammatory responses and defense systems, (2) oncometabolites, unexpected products from altered metabolism that are involved in tumorigenesis, (3) damaged metabolites, chemically-reactive compounds resulting from enzyme errors or spontaneous reactions that are normally regulated by damage-control systems, (4) microbiota metabolites, metabolites secreted by gut microbiota affecting the host physiology, and (5) phytochemicals, the plant specialized metabolites exerting various bioactivities on human metabolisms.
Therefore, metabolomics is an attractive research field, offering new biological insights to small molecules while there are still outstanding issues, especially in “informatics” to provide wider coverage of metabolomes.
Mass spectrometry is the popular platform in metabolomics to measure the small molecules by ionizing them, and the compound structure can be determined (mainly) by checking the “mass spectrum” pattern generated from the “mass fragmentation” of the ionized metabolite. Mass fragmentation occurs by adding energy (called collision energy) to a small molecule. The fragmentation scheme is very specific to the compound structure, enabling us to identify the unique metabolites in biological samples.
However, a long-term, outstanding issue in mass spectrometry-based metabolomics is that the complete prediction of mass fragmentation is not yet achieved, and, therefore, the identification of small molecules still depends on the confirmation of authentic standard compounds. The coverage of traceable metabolites is still limited to around 1,000 due to the lack of standard compounds, while it is believed that the small biomolecule world exceeds one million chemical species. Therefore, the next stage of metabolomics is to decode the physical/chemical phenomena of ionized metabolites as well as to handle the complicated “big data” from mass spectrometry. It’s no exaggeration to say that understanding mass fragmentations is linked to the deeper understanding of metabolisms.
To date, “computational mass spectrometry” is a growing research field assisting in the interpretation of mass fragmentations and elucidating unknown structures with metabolome databases and repositories. Here, two approaches, i.e. bottom-up and top-down approaches, are highlighted.
The feature of the bottom-up approach is to create the theoretical mass spectrum by extrapolating spectrum knowledge to structurally-similar or same-scaffold compounds (see figure). For example, lipids forming cellular membranes have a large variety of acyl chains in a lipid class, and the lipid diversity is involved in many physiological functions. In a mass spectrometer, the fragmentation scheme in a lipid class (e.g. phosphatidylcholine) is mostly universal in the acyl chain varieties: the ester bonds in higher polarity region are cleaved owing to the uneven distribution of collision energy in a molecule.
Once the fragmentation scheme of the lipid class has been interpreted, therefore, the diversity of certain lipid class can be grasped by creating the theoretical spectrum. This bottom-up approach is now expanded to wider metabolite classes, including oxidized phospholipids and plant specialized metabolites like phenylpropanoids, flavonoids, and glycoalkaloids, which are used to find alterations of lipid homeostasis linked to various diseases and to accelerate drug discoveries in natural product chemistry.
The feature of another approach, i.e. top-down approach, is to search reported molecular structures followed by a ranking of the structure candidates with evaluation techniques that untangle structure-spectrum relationships (see figure). Nowadays, we can utilize more than 200,000 mass spectral records from public repositories, and it can be used as a “training set” to construct fragmentation theory and machine learning for molecular structure predictions.
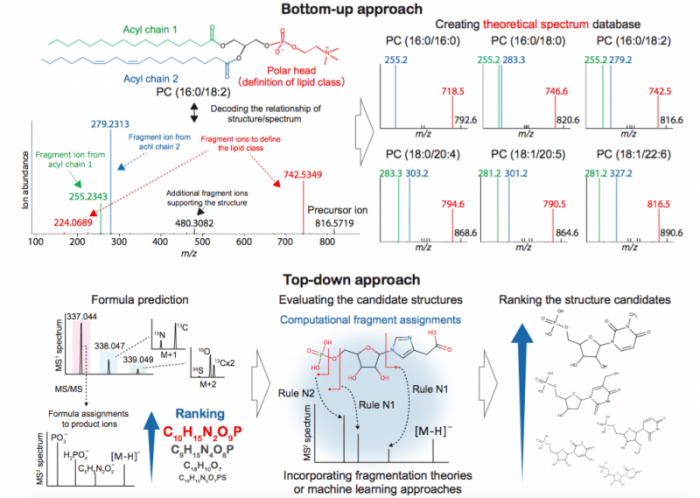
Figure 1: A bottom-up vs. top-down approach. Credit: Hiroshi Tsugawa
The accuracy of the top-down approach depends on the chemical spaces of interest, but the CASMI (critical assessment of small molecule identification) 2017 contest reported that the approach correctly assigned 37% (91/243), 61% (148/243), and 79% (193/243) of challenges as the top, top 3, and top 10 candidates, respectively.
This accuracy can be improved by database selections and curations in specific organs, tissues, and species. Especially in natural product research, taxonomical filters that apply information on species-chemical relationships efficiently exclude false-positive candidates. The practical use of the top-down approach is to narrow down the structure candidates finally determined by checking the authentic standards, and it can also enable us to identify novel modified metabolites removed from its canonical function in anabolism or catabolism by enzyme errors or chemical damage.
Improving bottom-up/top-down approaches and integrating these with additional approaches will facilitate the global identification of human, plant, and microbiota metabolomes. The final goal of computational mass spectrometry is the total understanding of mass fragmentation of small molecules, and for this purpose, updates on analytical chemistry with computational mass spectrometry are essential for the elucidation of new physiological function and biological mechanisms.
These findings are described in the article entitled, Advances in computational metabolomics and databases deepen the understanding of metabolisms, recently published in the journal Current Opinion in Biotechnology. This work was conducted by Hiroshi Tsugawa from the RIKEN Center for Sustainable Resource Science and the RIKEN Center for Integrative Medical Sciences.








