
We know DNA as the genetic blueprint of life. DNA can be chemically synthesized and used to build nanoscale structures and devices. DNA-based nanodevices can respond to cues such as other molecules, pH, temperature, and ionic conditions, and can also be used as biosensors.
Some DNA nanodevices are designed to provide visual outputs that tell us about the molecular events that occurred. For example, scientists have previously used atomic force microscopy (AFM) to display single nucleotide polymorphisms (a DNA sequence variation that causes abnormalities) as alphabetic characters and to compute simple math such as multiplication that is displayed on a nanoscale digital display. In my recent research, shape-changing DNA nanostructures are used to recognize and react to specific input DNA strands, with the resulting output graphically displayed using gel electrophoresis.
The DNA nanoswitch used in this study has two states. The “off” state is a linear duplex formed by a long single strand (~7 kilobases) hybridized to short complementary backbone oligonucleotides. Two of the backbone oligonucleotides can be modified to contain single-stranded extensions that are partially complementary to another DNA strand (the input data strand). Binding of the input strand to the nanoswitch creates a looped “on” state that can be easily identified on a gel (see figure).
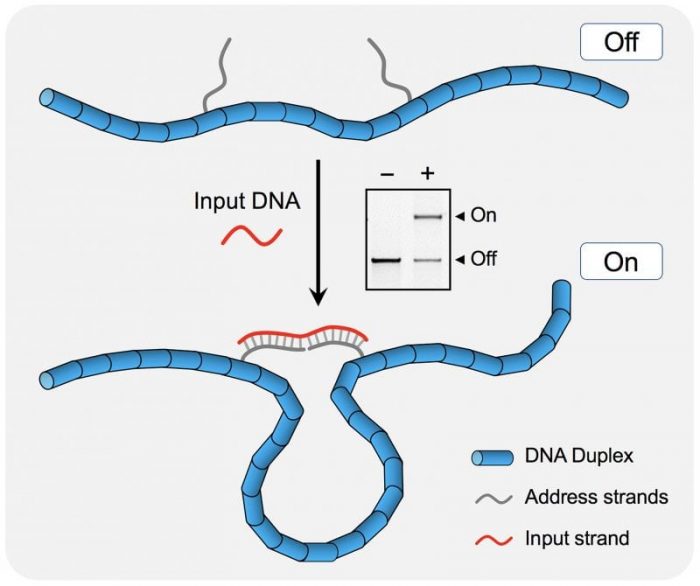
Copyright WileyVCH Verlag GmbH & Co. KGaA. Reproduced with permission.
The DNA nanoswitch is programmable. Address sites can be inserted anywhere along the scaffold thus resulting in different loop sizes (that migrate to different positions on a gel). Nanoswitches with multiple address sites are used to create “DNA pixels” to store and display alphanumeric characters. The read-out of the stored information is inspired by digital display circuits where a series of bits display a particular pattern (for example, a number or a letter). This multi-input nanoswitch can recognize different input molecules. These input DNA strands are complementary (i.e. can bind) to a pair of address sites (see figure). Addition of these input DNA strands trigger loops of specific sizes, which result in different bands on a gel.
Now imagine the gel as a segmented display board with a 5×5 readout matrix, where each looped band provides a “pixel” on the matrix. As an example, specific input strands were used to trigger and display three alphabet characters “r”, “N”, and “A” (see figure below). While this is not a pixelated image display such as the recent Mona Lisa made out of DNA origami plates, the main use of this system lies in a simple digital output for molecular reactions.
Copyright WileyVCH Verlag GmbH & Co. KGaA. Reproduced with permission.
We had earlier encoded the words, “Hello, world,” using this system, erased it using toehold-mediated strand displacement (where the input strand is removed using a fully complementary DNA strand), and rewrote, “Good bye,” on the same system. A library of such nanoswitches with distinct address strand pairs can be the future of point-of-care biosensors and DNA barcodes.
These findings are described in the article entitled Reconfigurable DNA nanoswitches for graphical readout of molecular signals, recently published in the journal ChemBioChem. The original research described in this article was performed by Arun Richard Chandrasekaran from Confer Health, Inc.









