
Amyloids are fibrillar proteinaceous assemblies whose accumulation is a hallmark of several human diseases. Formation of amyloid fibrils occurs when protein monomers change their structure and clump together. This kind of irreversible polymerization can lead to the accumulation and propagation of toxic oligomeric seeds. Such accumulation of amyloid fibrils takes place in more than 20 distinct diseases, including Alzheimer’s disease and type II diabetes.
Conversely, the last decade has witnessed the discovery of a second class of amyloids that are key to sustain life; these are the so-called functional amyloids and, in contrast to pathological amyloids, these fibrils readily disassemble after carrying out their functions.
Among the emerging number of functional amyloids uncovered to date, perhaps the RIPK1-RIPK3 necrosome core is the most perplexing. This is the first example of a human signaling complex of amyloidal nature. More precisely, the association of these two proteins RIPK1 and RIPK3 through their so-called RIP homotypic interaction regions (RHIMs), enables the formation of a functional amyloid fibril that serves to signal in a programmed cell death known as necroptosis. In contrast to all amyloid fibrils reported to date, the RIPK1-RIPK3 necrosome amyloid fibrils build on the assembly of two distinct proteins, instead of one protein. That is, this is the first example of a hybrid amyloid. By this unique feature, the RIPK1-RIPK3 necrosome has revolutionized the amyloid paradigm. Therefore, it is crucial to understand how the incorporation of two distinct proteins is favored over the homo-oligomerization process of protein self-stacking during amyloid formation.
The development of new solid-state Nuclear Magnetic Resonance (ssNMR) method has proven a powerful tool to elucidate amyloid structures. Therefore, the authors used ssNMR to unravel the enigmatic structure of the necrosome core formed by RIPK1 and RIPK3 at atomic resolution. The samples used for the structural study were obtained using molecular biology tools. Briefly, the strategy consists of feeding E. coli bacteria with 13C- and 15N-isotopically labeled material as the carbon and nitrogen sources for cell growth. By this method, it is possible to force bacteria cells to produce the proteins of interest (RIPK1 and RIPK3) in large amounts. Carbon and nitrogen atoms in these proteins will be enriched in 13C- and 15N isotopes, which are visible to the NMR technique. Proceeding this way, the authors isolated hybrid amyloid fibrils formed by RIPK1 (amino acid residues 496-583) and RIPK3 (residues 388-518) that were subjected to ssNMR analysis.
The interpretation of the solid-state NMR data is like solving a puzzle. Just like puzzle pieces need to come together to the right place to provide the right picture, the NMR data affords a large number of distances between a pair of atoms. There is only one way in which all such distances simultaneously fit a structural representation. In the case of the RIPK1-RIPK3 amyloid fibrils, the authors were able to obtain a number of distances below 8 Å between amino acid residues in the two proteins, which ultimately yielded the 3D structure of the amyloid complex at high resolution.
The structure of the RIPK1-RIPK3 illustrates for the first time the basis for hetero- over homo-oligomeric assembly, or how distinct proteins preferentially stack with each other rather than with themselves. It is interesting that among the ~90 and ~130 residues contributed by RIPK1 and RIPK3, respectively, only a restricted number is required for hetero-amyloid assembly or hybrid amyloid formation. Figure 1 illustrates how the two amyloidal, core sequences of both proteins align to allow protein alternation.
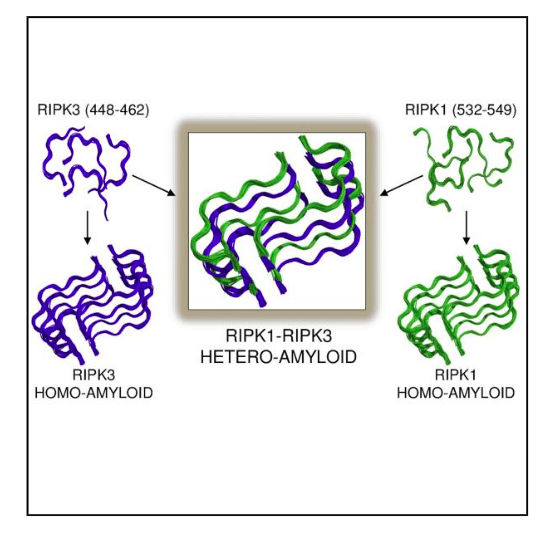
Figure 1: Illustrating how few amino acid residues in RIPK3(blue) and RIPK1 (green) drive homo-amyloid formation incorrect activation with analogous architecture to those of the more favorable hetero-amyloid determined by ssNMR. Republished with permission from Elsevier from https://doi.org/10.1016/j.cell.2018.03.032.
The striking feature of the core structure is how stacking with essentially identical conformations can form hybrid amyloid fibrils despite differences in amino acid composition. Whereas peptides bearing the core RHIM sequences can each form homo-amyloids themselves with the same architecture observed in the RIPK1-RIPK3 hetero-amyloid (Figure 1), the ssNMR structure of this hybrid amyloid provides a potential explanation for this preference; namely, the protein monomers in the fibrils are more tightly bound by alternately stacking RIPK1, RIPK3, RIPK1, RIPK3, etc. with respect to both the homo-RIPK1 and homo-RIPK3 counterparts.
This work is significant and has broad implications in the structural and human biology of cell death and beyond. The RIPK1-RIPK3 core is the first detailed structural study of a hetero-amyloid at atomic resolution, therefore providing a structural basis for understanding the mechanisms of signal transduction and hetero-amyloid formation in broader contexts. The first step towards rational drug design strictly requires the determination of the 3D structure of the target protein. Elucidating the key structure of the RIPK1-RIPK3 core could pave the way for the development of inhibitors of the necrosome core, whose incorrect activation is involved in some complex pathologies.
These findings are described in the article entitled The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex, recently published in the journal Cell. This work was conducted by Miguel Mompeán, Wenbo Li, Jixi Li, Ségolène Laage, Ansgar B. Siemer, Gunes Bozkurt, Hao Wu, and Ann E. McDermott from Columbia University, Harvard Medical School, and Boston Children’s Hospital.








