With the rise in bacterial strains that are resistant to available antibiotics, there is a growing need to better understand how gene function can influence the sensitivity to antibiotics, as well as to identify potential new drug targets. To this end, bacterial cell imaging is used to directly observe how chemical or genetic interventions alter growth and morphology at the single-cell level over extended time periods.
While static images of individual cells can be informative, additional insights can be gained by assessing how bacteria respond to a range of concentrations of chemicals. For example, antimicrobials can be assessed for their ability to alter cell morphology prior to inhibiting growth. In addition, the consequences of tuning gene expression over a range of transcriptional levels on cell growth and morphology can be tested by image analysis of cells throughout a concentration gradient.
To develop a system that incorporates the above distinct features in a cost-effective platform, we developed a microscale chip designed and built using adhesive tapes and a transparency sheet. This approach offers the advantage of not requiring expensive polymers or lithography fabrication steps typical of more conventional microfluidics systems, while still allowing high-resolution detection of single bacterial cells. The stacked layout of the chip incorporates a thin agar gel membrane over which bacterial cells are cultured. Thin conduits allow us to establish concentration gradients of pre-specified chemicals and drugs in the agar gel membrane. An environmental chamber with an upright benchtop microscope is designed to culture the bacterial cells, along with an autofocusing hardware/software module to adjust the microscope’s focus periodically. With the current setup, we can record both single bacterial cells and their populations over 10 hours automatically with no manual intervention. The hardware used here is low-cost and custom-built using off-the-shelf components and materials.
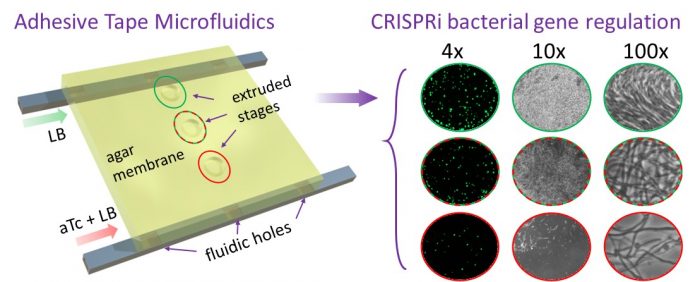
Figure reproduced with permission from ACS from https://pubs.acs.org/doi/full/10.1021/acssensors.9b01031
Bacterial cells were cultured in the platform and exposed to known concentration gradients of ampicillin with single-cell recording for over 10 hours. Morphological changes in the cells at different levels of the cell division inhibitor ampicillin were observed from the video recordings, including cell elongation, bulging, spheroplast formation, and eventual cell lysis.
We next incorporated CRISPR-interference (CRISPRi) into the system to allow direct visualization of cells as they respond over a range of transcriptional activity. For this, we engineered an E. coli strain to regulate expression of the essential ftsZ cell division gene by CRISPRi. By establishing a gradient of the inducer anhydrotetracycline (aTc), we observed a range of morphological responses to depletion of the FtsZ protein, as evidenced by increasing cell length and eventual cell death in response to an increasing concentration of the inducer. We were also able to determine the minimal inducer concentrations necessary to achieve discrete morphological changes in a single experiment.
In our opinion, the ability to control gene expression over a wide range using CRISPRi and measure the resulting responses of individual cells over time offers new opportunities to investigate and overcome resistance mechanisms, including testing the responses to novel antimicrobials and identifying new drug targets through knockdown of gene expression. The ability to monitor cell growth over extended time periods also allows investigation of antibiotic tolerance and persistence, which also can contribute to antibiotic treatment failure.









