Bacteria have been around for over a billion years and have developed remarkable capabilities to not only cause deadly infections but also to survive adverse environmental conditions (1, 2). This has led to a rich cellular diaspora with diverse geno- and phenotypic variability that is important to characterize owing to its dire implications on mankind.
Researchers are constantly evolving strategies that allow them to study complex cellular processes like cell-cell interactions, cellular metabolic activity, gene function, response to external stimuli (3-7), host-pathogen interactions, and surface-associated redox activities (8) at a single cell level. The foremost requirements of these single cell test platforms are to distinguish, isolate, manipulate, immobilize, and characterize cells in an easy and reproducible manner.
The conventional methods widely used for single cell isolation include fluorescence- or magnetic-activated cell sorting (FACS/MACS), micromanipulation, microdissection, and manual cell picking. While FACS/MACS need high cellular loads, large sample volumes and specific treatment of cells prior to analysis, the remaining techniques have quite a low throughput and require highly-skilled labor for carrying out the experiments. Other methods for cell capture include ink-jet printing on molecularly-patterned templates (9) or dip-pen nanolithography (10), but they again, suffer from poor
reproducibility, high cost, and our inability to reuse the substrate.
With recent advances in the field of microfluidics, manipulation and separation of cells on the basis of their dielectric properties, using external electric fields has garnered significant attention. This approach offers numerous advantages such as a high extent of miniaturization, low sample volume, controlled fluid dynamics, ease of sample handling, high throughput, great precision, automation, and label-free operation.
Motivated by these factors, we exploited an electrokinetic phenomenon called alternating current (AC) dielectrophoresis to collect live bacterial cells into grid-like patterns, with each node containing a single cell (see image below). This electric field-assisted assembly not only provided high spatio-temporal control over several thousands of cells simultaneously, but the dynamically-formed cellular arrays were also fully reversible meaning that both the cells and the chips could be retrieved at the end of each capture cycle. This study was recently published in Biosensors and Bioelectronics (11, 2018, 159–165) and showed that model Gram-negative bacteria S.typhi and E.coli can be trapped inside micropatterned conductive circular well electrodes by applying AC electric fields (5 MHz, 5-20 Vpp) across a few microliters of cell suspension.
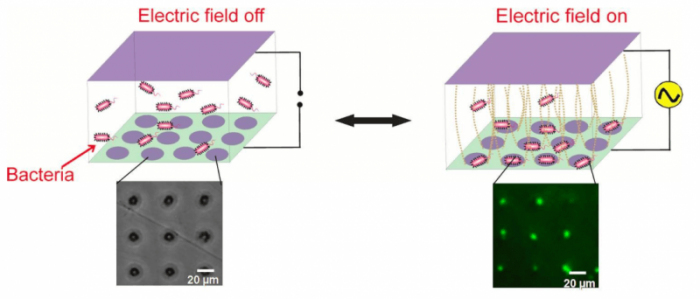
Dynamic electropatterning of live bacteria into single microwell arrays using AC electrokinetics. The bacteria were fluorescently tagged to allow visualization. Image courtesy Meenal Goel & Shalini Gupta
Shrinking the size of the individual electrodes down to 5 μm allowed single cells to be captured per well with 90% efficiency. This trapping of cells took place due to positive dielectrophoresis that attracted them toward regions of high electric field intensity (microwells in our case). The cells remained alive during the one-hour-long operation, and the overall collection efficiency (from bulk to the surface) was also found to be around 90%.
The potential biotechnological application of our chip was demonstrated in two ways. First, the two cell types were mixed in suspension and collected together using the chip. Counting their relative concentration in the 2D matrix using their phenotypic traits allowed us to directly estimate cellular bulk concentrations in the mixture without generating any calibration curves. Both the total time taken and the volume of the sample used was a fraction of the conventional methods. In the second case, the chip was integrated with an impedance spectroscope to carry out rapid cell viability testing. The response of dielectrophoretically-trapped bacteria against a known antimicrobial peptide was recorded at different drug concentrations and found to be ultrasensitive, reducing the overall time for drug susceptibility testing to just under an hour. This is a significant reduction over standard clinical methods that take several hours to produce outcomes.
In the future, our technology could be used for a wide range of applications including multiplexed sensing, rapid single bacterial profiling in heterogeneous populations, and ultrafast drug susceptibility testing to reduce the use of broad-spectrum antibiotics and, hence, the emergence of new antibiotic-resistant strains.
These findings are described in the article entitled Electric-field driven assembly of live bacterial cell microarrays for rapid phenotypic assessment and cell viability testing, recently published in the journal Biosensors and Bioelectronics. This work was conducted by Meenal Goel, Abhishek Verma, and Shalini Gupta from the Indian Institute of Technology Delhi.
References:
- Turner, N. A., Harris, J., Russell, A. D., Lloyd. D. 2000. J. Appl. Microbiol. 89, 751–759.
- Davey, M. E., O’Toole, G. A., 2000. Microbiol. Mol. Biol. Rev. 64, 847–867.
- Flaim, C.J., Chien, S., Bhatia, S.N., 2005. Nat. Methods 2, 119–125.
- Hung, P.J., Lee, P.J., Sabounchi, P., Lin, R., Lee, L.P., 2005. Biotechnol. Bioeng. 89, 1–8.
- Lee, M.Y., Dordick, J.S., 2006. Curr. Opin. Biotechnol. 17, 619-27.
- van der Meer, J.R., Belkin, S. 2010. Nat. Rev. Microbiol. 8, 511–522.
- Ziauddin, J., Sabatini, D.M., 2001. Nature 411, 107–10.
- Potma, E. O., de Boeij, W. P., van Haastert, P. J. M., Wiersma., D. A. 2001. Proc. Natl. Acad. Sci. 98, 1577–1582.
- Xu, Luping, Robert, Lydia, Ouyang, Qi, Taddei, François, Chen, Yong, Lindner, Ariel B., Baigl, Damien, 2007. Nano Lett. 7, 2068–2072.
- Kim, J., Shin, Y.-H., Yun, S.-H., Choi, D.-S., Nam, J.-H., Kim, S.R., Moon, S.-K., Chung, B.H., Lee, J.-H., Kim, J.-H., Kim, K.-Y., Kim, K.-M., Lim, J.-H., 2012. J. Am. Chem. Soc. 134, 16500–16503.








