
In the human genome, genetic information is carried on two copies (two chromosomes), each copy being inherited from one of the parents. For most genes, both copies (maternal and paternal) are expressed equally. However, it has been shown that ~150 human genes have been expressed from either the paternal or the maternal chromosome but not from both. These genes are called “imprinted genes” and are subject to “genomic imprinting.”
Genomic imprinting is a physiological process defined as the monoallelic expression of a gene according to its parental origin. A gene becomes imprinted when special epigenetic markers are attached to specific areas around it (without affecting the genomic sequence). These epigenetic marks can turn genes off or on; thus, playing key roles in cognitive development, metabolism and growth, especially fetal growth.
Genomic imprinting reflects a conflict between the mother and the father regarding fetal growth, without requiring a change within the genetic coding. On the one hand, the mother needs to survive the pregnancy and breastfeeding period; thus, she needs a fetus who does not deplete all her energy stores, who is not too large to deliver vaginally, and who does not require too much breast milk to survive. On the other hand, the father needs his progeny to survive and pass on his genes; therefore, he needs a large fetus and a big, strong newborn who would grow into a big, healthy child to spread his genes to the next generation as an adult. This conflict is also reflected by the nature of the genes expressed from each parent; paternally expressed genes (PEGs), usually coding functional proteins, are known to enhance fetal and post-natal growth, while maternally expressed genes (MEGs), usually not coding for proteins and kept at an intermediate level called non-coding RNA (ncRNA), are known to repress fetal and post-natal growth.
Imprinting disorders (IDs), caused by disturbances of imprinted genes, are a group of congenital diseases affecting growth, development, and metabolism in humans, leading to diseases with overlapping features, regardless of the genomic region affected. Temple syndrome (TS14, caused by imprinting disturbances on chromosome 14) and Silver-Russell syndrome (SRS, caused by loss of PEGs from chromosome 11) represent a particularly demonstrative example of clinical overlap between IDs. Both these syndromes include fetal and postnatal growth retardation, early feeding difficulties, early puberty, an increase in the risk of metabolic disorders and common physical traits. Moreover, TS14 patients also have a number of clinical features in common with another ID, Prader-Willi syndrome (PWS), a differential diagnosis for TS14, which is usually caused by loss of PEGs from chromosome 15 (Figure 1).
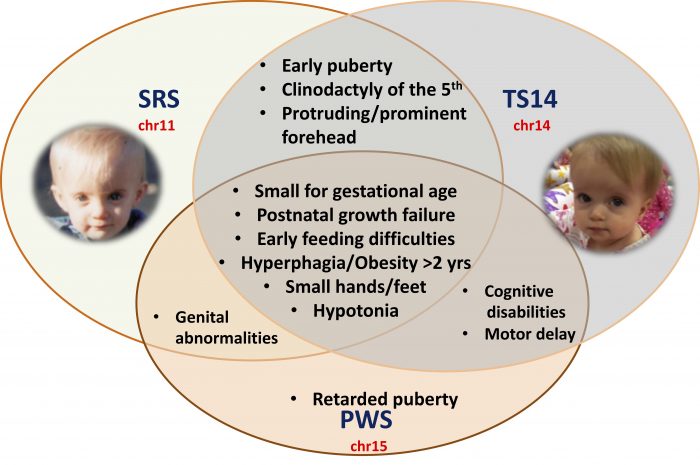
Figure 1: Schematic representation of the overlapping clinical features in SRS, TS14, and PWS patients*. Image republished with permission from Science Advances from https://advances.sciencemag.org/content/5/2/eaau9425 (* authorization obtained from the patient or the guardians to show the pictures)
To understand how TS14 patients, with imprinting defects on chromosome 14, present overlapping clinical features with SRS and PWS patients (with imprinting defects on chromosomes 11 and 15, respectively), we studied the expression of imprinted genes from chromosomes 14, 11 and 15 in TS14 patients. Interestingly, we found that TS14 patients i) lose expression of protein-coding PEGs and reactivate the expression of MEGs (ncRNA) from the paternal allele on chromosome 14; ii) lose expression of PEGs on chromosomes 11 and 15, as observed in patients with SRS and PWS; and, iii) display a large number of deregulated nonimprinted genes, half of which are commonly deregulated in SRS patients.
By using experimental designs to over-express ncRNA from chromosome 14 in normal cells, we were able to induce the loss of expression of imprinted genes on chromosomes 11 and 15, as seen in TS14 patients. These findings highlight the role of chromosome 14 ncRNAs as key regulators of the cross-talk between chromosome 14 and chromosomes 11 and 15 that may account for the clinical overlap between the three syndromes (Figure 2).
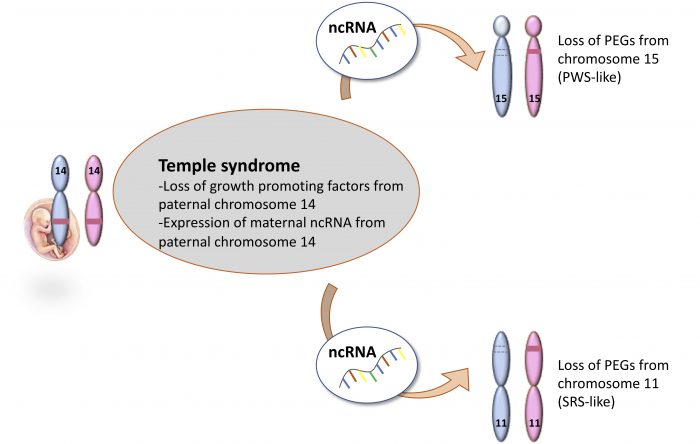
Figure 2: Schematic representation of the molecular findings and the mechanism from this study. Credit: author
In conclusion, we describe a concept of the interrelationship between the function and expression of imprinted genes, which is important not only for the diagnosis and care of patients with these rare disorders but also for a better understanding of fetal growth physiology.
These findings are described in the article entitled Transcriptional profiling at the DLK1/MEG3 domain explains clinical overlap between imprinting disorders, recently published in the journal Science Advances.









