Living cells rely on the transmembrane difference of electrochemical potential to store the energy needed for their biochemical reactions. Membrane potential is generated by respiratory chain proteins, which use the energy released during the oxidation of energy-rich substrates to translocate ions across the membrane against their electrochemical gradient.
The transmembrane difference in electrochemical potential is later used by cells to synthesize ATP, transport essential nutrients across the membrane, or energize cell flagella. The work of respiratory chains is central to the energetic metabolism of all living cells, and investigating their structure/function is key for understanding one of the most fundamental mechanisms in nature: energy transduction.
The promiscuity and protein composition of respiratory chains from different organisms is as diverse as life on Earth. Organisms are able to strive in the most variable conditions (e.g. sulfide-rich benthic regions, oxygen abundant ecosystems, or fertile nitrate subsoils) due to their respiratory chain composition. Humans, for example, are oxygen-dependent organisms since the final step in their respiratory chain is the reduction of oxygen to water (aerobic respiration). Different respiratory proteins are present in different taxonomic clades and attribute different respiratory abilities to cells.
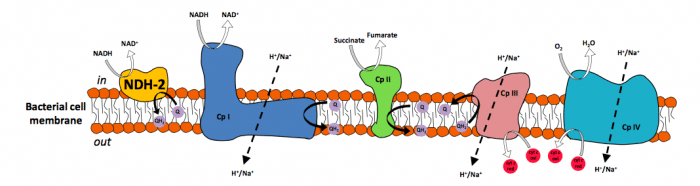
Type 2 NADH:quinone oxidoreductase (NDH-2), or alternative NADH dehydrogenase, is a membrane-bound respiratory protein which catalyzes the electron transfer from NADH to a quinone. NDH-2s are crucial for the catabolic metabolism since they contribute to the maintenance of the intracellular NADH/NAD+ ratio. They are also the only respiratory enzymes with NADH dehydrogenase activity present in the membranes of several pathogenic bacteria and protists. For this reason and for being considered absent in mammals, NDH‐2s were proposed as suitable targets for novel antimicrobial therapies. NDH-2 is considered a functional replacement to Complex I and a potential therapeutic resource for human neurodegenerative diseases, caused by Complex I failures (Parkinson’s disease and aging) since its expression restores the mitochondrial activity in animals with Complex I deficiency.
Structurally NDH-2s are divided into three domains: one for NADH (electron donor) binding, a second for the binding of FAD (its flavin cofactor), and a third domain responsible for the quinone (electron acceptor) binding and membrane interaction. Recent advances in structural biology allowed for the determination of the crystallographic structure of NDH-2s from different organisms, which re-opened the investigation race about this enzyme. X-ray structures from four different organisms have been recently determined, including the human pathogen Staphylococcus aureus (S. aureus).
The NDH-2 from S. aureus has been proposed as a suitable target for rational drug design, against this pathogenic organism. The enzyme from this organism is one of the most studied representatives from this protein family with previous studies focusing on its structure. Dr. Manuela Pereira and coworkers have determined the number and location of substrate binding sites and some general details about the enzymatic mechanism of NDH-2. The gathered information actively contributed to the understanding of this enzyme in other organisms as well as to the energetic metabolism of this organism.
The most recent study on S. aureus NDH-2 focuses on the molecular aspects of the enzymatic mechanism, specifically trying to unravel the role of the charge transfer complex (CTC). CTC is an electron sharing mechanism, which in the case of NDH-2 is formed between the nicotinamide ring of the substrate NADH and the alloxazine ring of the cofactor FAD upon substrate binding. The previously identified CTC manifests by an increase in the absorbance at 670 nm region, which allowed the authors of the study to use a set of spectroscopic techniques to study this biochemical formation. Using a combination of enzyme kinetics and UV-Visible spectroscopy, and using both physiological and nonphysiological electron acceptors (with distinct chemical properties) combined with established NDH-2 inhibitor (HQNO), the authors studied the CTC role in the two half-reactions of the enzyme mechanism.
The authors showed that the CTC slowed down the second half reaction of NDH-2 mechanism and proposed this to be a physiological mechanism of “keeping a low quinol/quinone ratio and thus avoid the production of radical oxygen species by unspecific oxidation of quinol.” The authors also associated for the first time the CTC formation to specific conserved structural features present in this enzyme family.
These findings are described in the article entitled Regulation of the mechanism of Type-II NADH: Quinone oxidoreductase from S. aureus, recently published in the journal Redox Biology. The work was led by Manuela Pereira from the University of Lisbon and represents the most recent contribution from the group about this protein and is an integrant part of the PhD projects from F. Sena and F. Sousa.









