
From animals to microorganisms, many species use sex pheromones to attract a potential partner of the opposite sex. The specificity of a pheromone and its corresponding receptor determines the ability of males and females to recognize each other; therefore, mutational alterations in the pheromone system can prevent mating. This so-called reproductive isolation, which restricts gene flow between populations, is considered a key evolutionary process, but how do new combinations of pheromone and receptor evolve?
Our recent study published in the open-access journal PLoS Biology demonstrated that an “asymmetric” pheromone recognition system in the fission yeast Schizosaccharomyces pombe: one pheromone/receptor pair operates extremely stringently, whereas the other pair is free to undergo a certain degree of diversification, perhaps leading to reproductive isolation in S. pombe [1].
The two sexes (“Plus” and “Minus”) of S. pombe (Fig. 1) each secrete a small pheromone peptide (“P-factor” and “M-factor”), which binds to a corresponding receptor on the surface of cells of the opposite sex. We previously created a novel fission yeast strain pair of opposite sexes that behave like new species (i.e., they can mate with each other, but mate only very poorly with wild-type strains), by genetically altering the primary structure of both M-factor and its cognate receptor [2,3]. When the novel strain was co-cultured with wild-type strains in a test tube, the level of inter-species exchange of genetic information was extremely low.
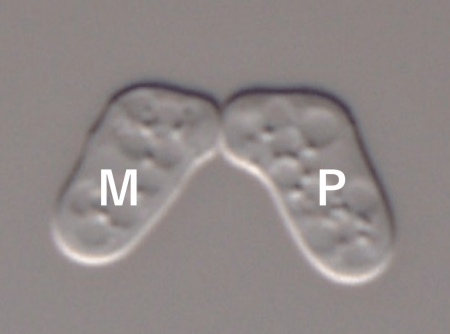
Fig. 1 Mating in S. pombe. The mating reaction between M- and P-cells requires both recognition of P-factor by its receptor and recognition of M-factor by its receptor. Image courtesy Taisuke Seike.
This experimental result supports the idea that alterations in pheromone/receptor recognition may be one of the mechanisms underlying reproductive isolation. More generally, however, loss of pheromone activity may result in the extinction of a species lineage, but how would a pheromone system evolve while maintaining successful mating in nature?
To answer this question, we recently explored similarities and differences between genes encoding the two pheromones and their receptors in 150 wild S. pombe strains of more than 22 different geographical origins (Fig. 2). Sequencing analysis clearly demonstrated that there is no variation in the amino acid sequences of one pair, the M-factor and its receptor, while the other pair, the P-factor and its receptor, show substantial diversity. Interestingly, such “asymmetric” diversification of the two pheromones was also seen in the closely related two fission yeast species S. cryophilus and S. octosporus, implying that communication by M-factor plays an important role in defining the species, rather than signaling by P-factor in fission yeasts.
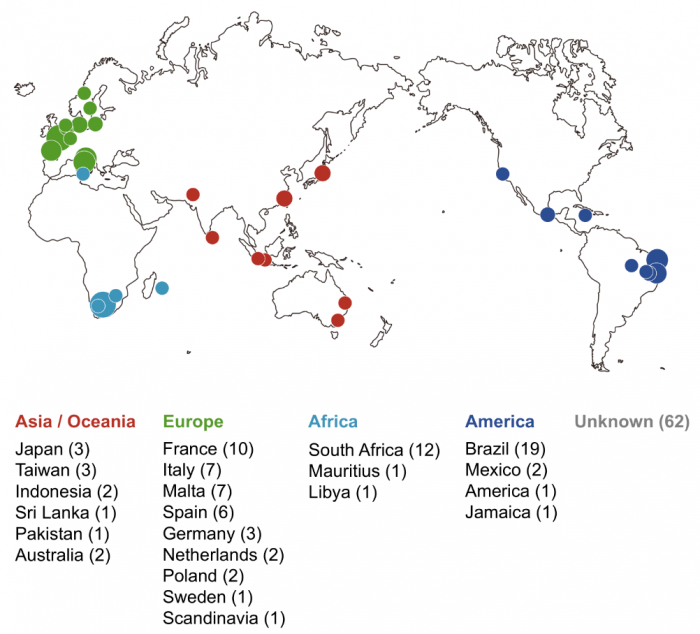
Fig. 2 Geographical origins of the 150 wild S. pombe strains. Colored circles indicate the approximate countries and regions of the isolated strains, which are color-coded by continents: Asia/Oceania, Europe, Africa, and America. Circle size increases in proportion to the number of strains isolated from each region; the actual number of strains is shown in parentheses. Republished from PLOS open access article https://doi.org/10.1371/journal.pbio.3000101
We found that S. octosporus M-factor does not stimulate a mating response from wild-type S. pombe, but the introduction of S. octosporus P-factor could partially restore physiological mating (approximately half of wild-type efficiency) in a laboratory S. pombe strain lacking its own P-factor gene. Similarly, the introduction of Sp-P-factor could partially restore mating in a laboratory S. octosporus strain lacking its own P-factor gene. Indeed, most of the P-factor peptide variants had physiological effects between S. pombe and S. octosporus, whereas M-factor peptide was dysfunctional in other species. In other words, M-factor operates only within the same species, whereas P-factor operates beyond species. These results suggest that two pheromone receptors might have distinct specificities for their corresponding pheromone.
At present, it remains unclear why S. pombe possesses an “asymmetric” pheromone recognition system (Fig. 3), but it might allow flexible adaptation to mutational changes in pheromones/receptors while maintaining stringent recognition of mating partners, perhaps leading to reproductive isolation.
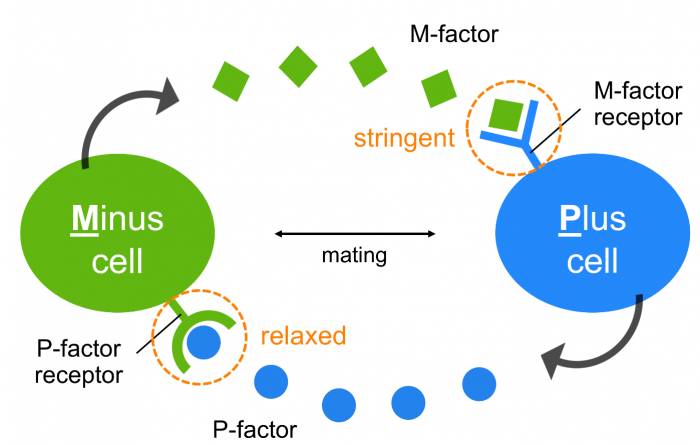
Fig. 3 An asymmetric system of pheromone recognition in fission yeast. The specificity of M-factor is extremely stringent, whereas that of the P-factor is relatively relaxed; therefore, the diversity of the two pheromones might facilitate asymmetrically in nature, perhaps as a first step toward reproductive isolation in fission yeast. Republished from PLOS open access article https://doi.org/10.1371/journal.pbio.3000101
Our findings contribute new insights into the evolutionary mechanisms. We speculate that many species might have similar systems for creating new versions of pheromones, allowing them to persist enough long in a population to evolve adaptations of receptors. Before a mutant is completely lost, a second suppressor mutation may occur to recover the first defect. Thus, the combination of a pheromone and its receptor might evolve step by step.
These findings are described in the article entitled Asymmetric diversification of mating pheromones in fission yeast, recently published in the journal PLOS Biology.
References:
- Seike T, Shimoda C, Niki H. (2019) Asymmetric diversification of mating pheromones in fission yeast., PLoS Biol., 17(1): e3000101.
- Seike T, Yamagishi Y, Iio H, Nakamura T, Shimoda C. (2012) Remarkably simple sequence requirement of the M-factor pheromone of Schizosaccharomyces pombe., Genetics, 191(3): 815–825.
- Seike T, Nakamura T, Shimoda C. (2015) Molecular coevolution of a sex pheromone and its receptor triggers reproductive isolation in Schizosaccharomyces pombe., PNAS, 112(14): 4405–4410.









