
While no one wants to receive a cancer diagnosis, a number of treatments are now available, making the likelihood of a favorable outcome better than ever before. A particularly attractive class of treatment is immunotherapy — a strategy that leverages a person’s own immune system to fight the cancer. Immunotherapy is often accomplished with oral medication, making it one of the more convenient ways to address this illness.
Among immunotherapies, many involve the generation of DNA damage.1 This may feel counterintuitive: as DNA is the fundamental biomolecule of genetic information for cellular function — the building block of life — it would not seem that damaging DNA would improve one’s health. Indeed, many forms of DNA damage are detrimental to health, and the body is equipped with a number of repair mechanisms to counteract this damage.2 However, certain types of DNA damage can trigger these immune responses to kill cancer cells. Many current therapies, such as cisplatin, methotrexate, and doxorubicin, leverage DNA damage, as do several emerging treatments.1
Given the emerging landscape of DNA damaging drugs, it would be highly beneficial to develop sensors that can follow drug activity to discover patient-specific responses. An ideal treatment would selectively target cancer while producing minimal side effects. However, it can be difficult to predict how a drug will impact a patient given the wide range of damage repair pathways in the body and the range of individual immune responses.3 To address this challenge, we propose to leverage a device to investigate the activity of drugs from patient samples to identify optimal treatments.
In our recent work with the Boothman Lab of the Indiana University School of Medicine, we demonstrated a means to follow DNA damage responses to anticancer drug treatment in lysates of cancerous and normal cells.4 We accomplished this with devices utilizing electrical transport within DNA.5 Gold electrodes patterned on chips are modified with forests of synthetic DNA bearing a probe molecule. Charge reaches the probe only if the DNA remains fully intact, but transport is lost if the DNA is structurally compromised by damage or damage repair responses. Cells were lysed — split open to create a solution of cellular contents — and added onto the chips modified with DNA. Anticancer drugs could then be added to this solution over the device, and damage responses recorded by the synthetic DNA on the chip. Higher damage responses were indicative of higher anticancer drug activity.
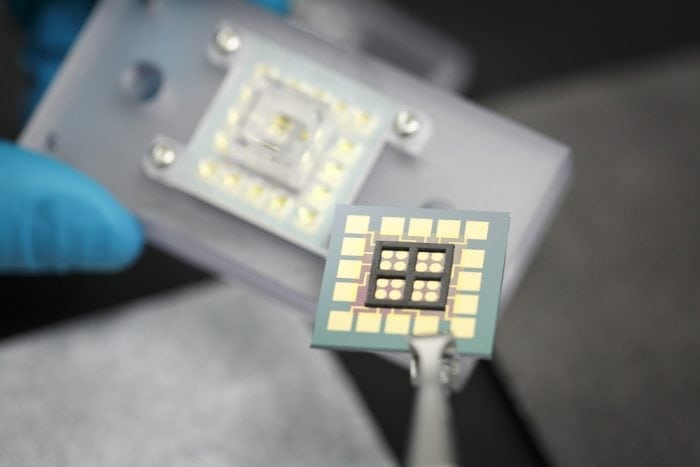
Credit: Jason D. Slinker
Using these devices, we showed that an experimental anticancer drug (clinical trial ARQ 761) exhibited approximately fourfold higher DNA damage in cancerous versus normal cells. If various aspects of the DNA damaging cycle of this drug were deactivated, it ceased to be effective in cancerous cells, demonstrating the mechanism of action. The dosage dependence of this drug was followed on the device and compared to cellular concentration responses. This revealed that DNA damage ensued at concentrations well below that needed for cell death and that high levels of damage were necessary to produce cell death.
We noticed, however, that we could not indiscriminately drop the lysates onto our chip. Rather, to accomplish this work, our devices had to be modified to match the biology of a typical cell. Device damage repair is accomplished by repair enzymes that are found in the nucleus with the DNA. Alternatively, the experimental drug is activated by enzymes outside the nucleus, in the cell cytoplasm, that generates hydrogen peroxide that diffuses into the nucleus and damages DNA. To reproduce the differences found in the cell, we separated nuclear and cytoplasmic lysates and maintained them in distinct wells over the chip that permitted solution interchange.
Overall, we envision that this technology could be leveraged to follow multiple anticancer treatments. That is, samples of cancerous and normal cells could be drawn from a patient, lysed, and added to our devices. Multiple DNA damaging immunotherapies could then be followed with our technology to determine which drug produces high damage in the cancer samples and low damage in the normal cells. This would simultaneously enhance drug effectiveness while minimizing side effects. Further studies will explore the generality of this approach and develop additional tools for assessing the impact of anticancer drugs.
These findings are described in the article entitled Following anticancer drug activity in cell lysates with DNA devices, recently published in the journal Biosensors and Bioelectronics. This work was conducted by Dimithree Kahanda and Jason D. Slinker from The University of Texas at Dallas, and Naveen Singh and David A. Boothman from Indiana University.
References:
- Cheung-Ong, K.; Giaever, G.; Nislow, C., DNA-Damaging Agents in Cancer Chemotherapy: Serendipity and Chemical Biology. Chemistry & Biology 2013, 20 (5), 648-659.
- Sancar, A.; Lindsey-Boltz, L. A.; Unsal-Kacmaz, K.; Linn, S., Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annual Review of Biochemistry 2004, 73, 39-85.
- Roos, W. P.; Thomas, A. D.; Kaina, B., DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2016, 16 (1), 20-33.
- Kahanda, D.; Chakrabarti, G.; McWilliams, M. A.; Boothman, D. A.; Slinker, J. D., Using DNA devices to track anticancer drug activity. Biosensors & Bioelectronics 2016, 80, 647-653.
- Genereux, J. C.; Barton, J. K., Mechanisms for DNA Charge Transport. Chem. Rev. 2010, 110 (3), 1642-1662.









