
Unconventional oil and gas development (UD) is the process of extracting oil and natural gas from relatively impermeable subsurface shale. Within UD, the process of hydraulic fracturing, more commonly known as ‘fracking’, is used to stimulate sequestered hydrocarbons from the target rock formations using a combination of water, sand, and chemical additives.
Throughout the course of its expansion across the United States, fracking has been one of the most controversial sub-topics in the climate change debate. Industry advocates have cited the vast economic benefits and the reduction of carbon emissions during the shale energy revolution, whereas skeptics are quick to list the potential environmental impacts. To this point, the scientific literature has indicated that UD and environmental stewardship are not mutually exclusive; however, a growing number of studies have also revealed the occurrence of groundwater contamination, surface spills, and toxic air emissions that are attributable to mechanical inefficiencies within the UD process.
More specifically, rogue natural gas, heavy metals, and chemical compounds have been detected in water wells located within close proximity to UD operations. It has not been until very recently that the scientific community in this field has started to examine the environmental implications of UD within the context of biological contaminants.
In three recently published studies, our team collected groundwater samples across the shale energy basins of Texas to examine the prevalence of harmful pathogens in clean and contaminated groundwaters. Our samples ranged from water wells with no reported issues and no detectable levels of deleterious metals and chemicals to water wells that had been contaminated by UD by either issue with faulty drilling infrastructure and/or errant well stimulation practices.
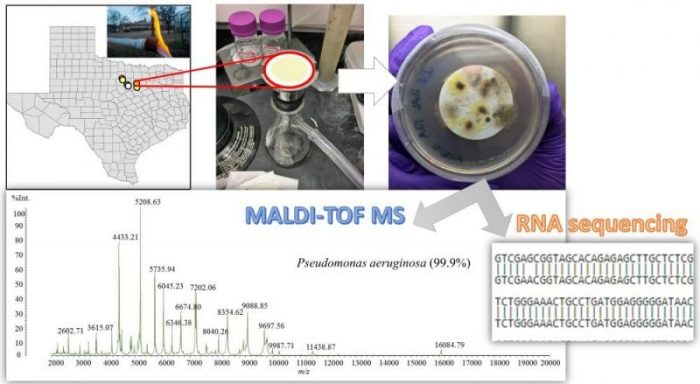
Credit: Zacariah Hildenbrand
The results were quite surprising. Not only did we find that various opportunistic pathogens could survive in the presence of hydrocarbon gases and chemical additives, they appeared to thrive and exhibited robust resistance profiles to multiple antibiotics. We even observed that certain pathogens were resilient to high levels of chlorination.
The presence of bacteria in groundwater is not a new phenomenon. Nevertheless, the breadth of species that are covered by the federal and state drinking water standards is very limited. Our novel methods using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) enabled us to screen the collected samples for over 2,500 different bacterial species simultaneously. As such, our detections of Pseudomonas aeruginosa, a bacterium that can deteriorate skin integrity upon contact, would previously go undetected by standard testing requirements.
The silver lining in this research is that two bacteria were identified in contaminated water wells that could be used as tools to biologically remediate chemicals from wastewater. Pseudomonas stutzeri and Acinetobacter haemolyticus both exhibited the ability to degrade chloroform and toluene under laboratory conditions, which is interesting given that they were isolated from water wells afflicted with the same chemicals. Collectively, these studies illustrate that various species of bacteria can adapt and thrive under contamination conditions, which adds another layer of complexity for deciphering how to handle instances of subsurface contamination.
The studies, Exploring the links between groundwater quality and bacterial communities near oil and gas extraction activities, and Characterization of bacterial diversity in contaminated groundwater using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry were published in the journal Science of the Total Environment. The article, MALDI-TOF MS for the identification of cultivable organic-degrading bacteria in contaminated groundwater near unconventional natural gas extraction sites was published in the journal Microorganisms. The work was performed by researchers and affiliates with the Collaborative Laboratories for Environmental Analysis and Remediation at the University of Texas at Arlington.









