Have you ever seen one of your genes? The specific part of your genome where it is found? I bet you haven’t, but neither have I! It’s not possible to see genes with the naked eye, but even if you could, chromosomes (where genes are housed) are huge, so where would you look? Being able to see them would be incredibly neat, and it could allow researchers to make exciting discoveries and doctors to develop personalized treatments.
Take the HER2 gene in humans, for instance. We’ve come to learn that it is located on the right arm of chromosome 17 and that it’s an oncogene – a gene associated with cancer (Kallioniemi, et al. 1992). HER2 is one of the three most commonly mutated genes in breast cancer, along with the estrogen and progesterone receptors (BRCA1 and BRCA2). In 15 – 25% of breast cancer cases, the HER2 gene is overexpressed (abnormally high) due to an increase in the number of copies that a person has or mutations in the gene that allow it to be switched ‘on’ far too often (HER2 Status).
When doctors know that patients have a mutated form of HER2 (termed HER2+), they can specialize treatments that are more specific than broad-spectrum cancer-treatment drugs. Imagine if doctors could look at every cancer patient’s HER2 gene to see if they carry the mutated form or have an increase in HER2 copy number. That would be an amazing way to personalize medicine to patients who are HER2+. Fortunately, whereas genes are too small to be seen with the naked eye, there is a way to do the next best thing – visualize the genes using molecular techniques.
The story of how we visualize genes all started in the early 1960s, after the discovery of DNA, but before we knew anything about the specifics of cancer genetics. It was already known that all living things have chromosomes made of DNA, residing in the nuclei of cells, and that DNA is transcribed into RNA. DNA and RNA readily pair together, or hybridize, as they are both made of nucleic acids. Scientists had also discovered how to immobilize DNA and hybridize it with radioactively-labeled RNA, called a probe (Nygaard & Hall, 1963). The radioactive RNA could then be visualized by autoradiography, on a nitrocellulose membrane (think black and white instant film cameras). However, RNA visualization occurred outside of cells, not within cells where genes and DNA are actually located (or in situ), so no one had been able to determine where specific genes were located on chromosomes.
In the mid-1960s, a researcher named Joseph Gall tried to take autoradiography a step further by squashing cell nuclei on a slide and then hybridizing radioactive RNA with the DNA – that is, attempting to hybridize DNA in situ. It didn’t work, as the signal was too weak to detect. However, science is about persistence and improving methods! Some of Gall’s earlier work demonstrated that within the nuclei of the huge egg cells of frogs and salamanders were hundreds of nucleoli, regions that play several roles including making ribosomes, machines within cells which read RNA and create proteins (Gall, 1968). Ribosomes are made up of ribosomal RNA (rRNA) that are derived from ribosomal DNA (rDNA).
Gall had previously discovered that rDNA reside within nucleoli, and he realized that since there were hundreds of nucleoli, there were also hundreds of copies of the same sequence of rDNA. This meant that he should then be able to hybridize the rDNA and generate a large enough signal for detection. Eureka – there was a plan of action! In the late 1960s Gall and his graduate student, Mary Lou Pardue, tried in situ hybridization in the egg cells of the model species Xenopus (African clawed frog), and this time it worked (Gall & Pardue, 1969)!
To follow up on these results, Gall decided to look at mouse chromosomes. Mice, another model species, also have rDNA, but they also have satellite DNA, a region of repeating DNA which contains no genes. Whereas the Xenopus rDNA was known to exist in the nucleoli, no one knew where satellite DNA was located in the mouse genome. This was the perfect opportunity to show that in situ worked and locate where mouse satellite is located! Once the satellite DNA was isolated, it was radioactively-labeled and the attempt at hybridization was made in 1970. It was again successful, and the results showed that satellite DNA was housed in heterochromatin, or inactive DNA, on the ends of the chromosomes (Pardue & 1970).
In contrast, species of Drosophila contain heterochromatic satellite DNA near the centers of their chromosomes, and it totals up to 41% of all the DNA in typical diploid tissues, such as the brain (Ritossa et al, 1966). Salivary tissue from Drosophila contain polytene chromosomes, very large chromosomes that can be duplicated up to thousands of times. When the satellite DNA was separated from salivary and brain tissue, the polytene chromosomes of the salivary tissue appeared to have almost no satellite DNA compared to the diploid brain tissue. So it appeared that no satellite DNA was present, but was that actually the case? When in situ hybridization was used to visualize the heterochromatic regions of DNA in each tissue type, satellite DNA was indeed found! It was discovered that while the rest of the chromosomes are massively duplicated, satellite DNA is not duplicated; therefore, the copy number of the heterochromatin regions is the same between the tissue types, yet it is difficult to separate the satellite DNA since the total amount of DNA is so high in polytene tissues (Gall et al, 1971).
Over time these and other successes led to in situ becoming a hugely popular method (Bishop 2010; Jin & Lloyd 1997; Levsky & Singer 2003). With improvements to the protocol, it even became possible to hybridize genes found in low copy number. Radioactively-labeled probes were replaced with safer, fluorescently-labeled probes in the late 1970s and early 1980s (Langer-Safer et al, 1982; Rudkin & Stollar, 1977). The advent of fluorescent in situ hybridization (FISH) even made it possible to hybridize multiple genes at the same time (Reid et al, 1992).
Currently, in situ hybridization has a fantastic number of applications beyond determining where individual genes are located on specific chromosomes. For example, it has been used to determine gene expression patterns by the identification of active and inactive DNA within different tissue types and to identify bacterial species based on genes specific to them (Gall, 2016). Perhaps most importantly with respect to our health, in situ hybridization allows for the rapid detection of a multitude of infectious and genetic diseases. As of 2017, there are over 30,000 online journal articles with “in situ hybridization” in their titles alone!
Although breast cancer is rare in men (Male Breast Cancer), it’s the most commonly diagnosed and the second leading cause of deaths in women (Breast Cancer Facts). Following the early experiments where Gall was able to determine that copy number of heterochromatin remained the same in diploid and polytene tissues in 1971, it became possible to determine that copy number of HER2 differed between diploid and cancerous tissue in 1992 (Kallioniemi, et al. 1992). Importantly, as in situ was one of the first tests able to detect different types of breast cancer, it helped to open the door to personalized medicine.
Specific drugs, including Herceptin, Kadcyla, Perjeta, and Tykerb, work by specifically targeting activity of the HER2 protein in breast cancer and are much more effective on HER2+ cancers than other treatments (HER2 Status). Indeed, only three tests are currently used to diagnose HER2+ breast cancers, and one is still a FISH method built on Gall and Pardue’s work. Although the HER2 gene is too small to see, it is now possible to visualize it. Just imagine, though, a world without the persistence of Gall, Pardue, and many others using model genetic organisms such as Xenopus, mice, and Drosophila. It’s possible that all we’ve learned with in situ hybridization, and the medical advancements that were built upon that work, might never have been developed. Luckily, we are in a world where they did persist, and we are reaping the rewards thereof.
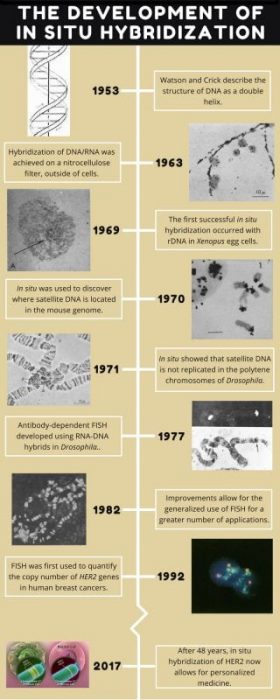
Image credit: Adam J. Ramsey
*For more information, check out Joseph Gall’s video on the development of in situ hybridization by the Carnegie Institute and this video with detailed information on HER2 and the action of Herceptin on HER2+ cancer cells.
This work is based on findings from research conducted by Adam Joseph Ramsey from the University of Memphis and as part of the Communication and Outreach Subcommittee of the Genetics Society of America.
References:
- Kallioniemi O, et al. 1992. ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proceedings of the National Academy of Sciences of the United States of America 89:5321–5.
- (2017, August 11). HER2 Status. Retrieved from http://www.breastcancer.org/symptoms/diagnosis/her2.
- Nygaard AP & Hall BD. 1963. A method for the detection of RNA-DNA complexes. Biochemical and Biophysical Research Communications. 12:98–108.
- Gall JG. 1968. Differential synthesis of the genes for ribosomal RNA during amphibian oögenesis. Proceedings of the National Academy of Sciences of the United States of America 60:553–560.
- Gall JG & Pardue ML. 1969. Formation and Detection of Rna-Dna Hybrid Molecules in Cytological Preparations. Proceedings of the National Academy of Sciences 63:378–383.
- Pardue ML & Gall JG. 1970. Chromosomal Localization of Mouse Satellite DNA. Science 168:1356–1358.
- Ritossa FM, Atwood KC, Lindsley DL & Spiegelman S. 1966. On the chromosomal distribution of DNA complementary to ribosomal and soluble RNA. National Cancer Institute Monographs. 23:449–72.
- Gall JG, Coren EH & Polan ML. 1971. Repetitive DNA Sequences in Drosophila. Chromosoma 33:319–344.
- Bishop R. 2010. Applications of fluorescence in situ hybridization (FISH) in detecting genetic aberrations of medical significance. Bioscience Horizons 3:85–95.
- Jin L & Lloyd RV. 1997. In situ hybridization: methods and applications. Journal of clinical laboratory analysis 11:2–9.
- Levsky JM. 2003. Fluorescence in situ hybridization: past, present and future. Journal of Cell Science 116:2833–2838.
- Langer-Safer PR, Levine M & Ward DC. 1982. Immunological method for mapping genes on Drosophila polytene chromosomes. Proceedings of the National Academy of Sciences of the United States of America 79:4381–5.
- Rudkin GT & Stollar BD. 1977. High resolution detection of DNA-RNA hybrids in situ by indirect immunofluorescence. Nature 265:472–473.
- Ried T, Baldini a, Rand TC & Ward DC. 1992. Simultaneous visualization of seven different DNA probes by in situ hybridization using combinatorial fluorescence and digital imaging microscopy. Proceedings of the National Academy of Sciences of the United States of America 89:1388–1392.
- Gall JG. 2016. The Origin of In Situ Hybridization – a Personal History. Methods 98:4–9.
- (2016). Male Breast Cancer. Retrieved from http://www.nationalbreastcancer.org/male-breast-cancer.
- (2016). Breast Cancer Facts. Retrieved from http://www.nationalbreastcancer.org/breast-cancer-facts.









