
In past decades, nucleic acid drugs have gained significant attention thanks to the tremendous potential for the treatment of inherited diseases. However, efficient and biocompatible delivery of nucleic acids still remains challenging.
Cationic lipids can pack nucleic acids into nanoparticles via electrostatic interaction, while the cytotoxicity and possibility of negatively-charged serum proteins binding at physiological pH greatly restrict further applications. Nucleoside-based lipids, which possess a lipid moiety and a highly specialized polar head-group (adenosine, thymidine, cytidine, guanosine, uracil, or their analogs), provide additional hydrogen bonding with complementary bases via Watson-Crick base pairing and π-stacking. In this paper, four kinds of nucleobase-lipids were prepared and named DXBAs (DOTA, DNTA, DOCA, and DNCA), and the assembly, biological properties, and functional mechanisms of DXBAs when interacting with nucleic acid drugs (oligonucleotides and plasmid DNAs) were systematically investigated.
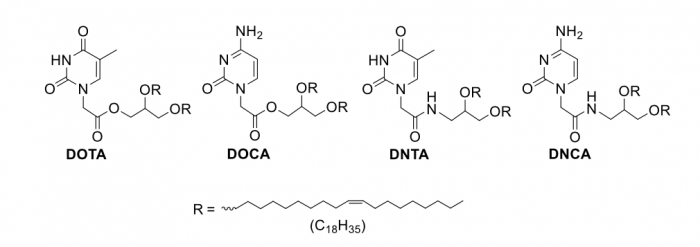
Figure 1. Chemical structures of nucleobase-lipids. Credit: Zhenjun Yang
All these nucleoside-based lipids (as well as their nucleic acid complexes) self-assemble for the formation of nanoparticles with 150 – 220 nm diameters. Interestingly, the DXBAs (especially for DNCA) mainly bind to single-stranded oligonucleotides rather than the duplexes. Further studies show that cytidine-based lipids (DNCA) can form more compact structures than thymine-based lipids, and amide linkage possesses better hydrophilic than an ester linkage. Thus, DNCA lipids exhibit the best self-assembly properties among DXBAs analogs.
Guanine-rich (G-rich) DNA and RNA molecules can fold into non-canonical structures stabilized by the stacking of G-quartet (G4), which is widely used in bioanalysis and biomedical applications. The G4-aptamer AS1411 encapsulated by DNCA may promote the formation of G-quartet structures in intracellular condition, and significantly enhance the permeation and antiproliferative ability. Besides, DNCA/AS1411 nanoparticles are rarely co-localized with lysosomes, suggesting related-enzymes hardly degradation of this nucleic acid drug.
Additionally, DNCA/AS1411 nanoparticles importantly stop the cell cycle at the S-phase and enhance the early apoptosis than naked AS1411s. Further, AS1411 (Cy7 labeled) delivered by DNCA remarkably prolong the drug duration after the treatment of BALB/c mice suffering A549 tumor by peritumoral injection. All data show that DNCA is a successful vehicle for AS1411 delivery and exerts biological activity improvement in vitro and in vivo.
The delivery efficiency of miRNA and antisense oligonucleotide, which silence or inhibit the target mRNA expression, were evaluated as well. As expected, the cellular uptake of ssRNAs was higher than dsRNA. Besides, the peptide-conjugation RNA specifically interacted with DNCA lipid and improved the nanoparticle stability. More importantly, DNCA and peptide-conjugation enable to regulate the endocytosis pathway, resulting in less lysosome degradation. Additionally, the antisense oligonucleotide can be delivered by DNCA lipid despite a low level of efficacy; however, high transfection efficiency can recover through doping with a slight of cationic lipid (CLD) in DNCA.
Plasmid DNAs are traditional nucleic acid drugs, which are frequently used in therapeutics. DNCA displays outstanding characteristics in serum-containing environments, with transfection efficiency better than that of Lipofectamine 2000. For plasmid DNA transfection, delivery efficiency of 10-20 μM DNCA is basically equivalent to that of 1 g∙L-1 Lipofectamine 2000. All data show that DNCA transfection has great potential for use with therapeutic G4 aptamers, as well as in other nucleic acid-based drug applications.
These findings are described in the article entitled Annealing novel nucleobase-lipids with oligonucleotides or plasmid DNA based on H-bonding or π-π interaction: Assemblies and transfections, recently published in the journal Biomaterials. This work was conducted by Yuan Ma, Yuejie Zhu, Chao Wang, Delin Pan, Shuang Liu, Mengyi Yang, Zhangping Xiao, Xiantao Yang, Wenting Zhao, Xinyang Zhou, Yiding Li, Yufei Pan, Jing Sun, Shuhe Wang, Zhu Guan, Lihe Zhang, and Zhenjun Yang from Peking University. This work was supported by the Ministry of Science and Technology of China (Grant No. 2012CB720604, 2017ZX09303013) and the National Natural Science Foundation of China (Grant No. 21778006, 21332010).









